













| General Information: |
|
| Name(s) found: |
MTR10_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 972 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNLQVSDIE TALQCISSTA SQDDKNKALQ FLEQFQRSTV AWSICNEILS KEDPTNALLE 60
61 LNIFAAQTLR NKVTYDLSQL ENNLPQFKDS LLTLLLSHNQ KLIITQLNVA LARLAIQFLE 120
121 WQNPIFEIIS LLNSSPSILL NFLRILPEET LDIASTSLTE VEFNSRIHEL IDPIAEDVLK 180
181 FLVSCIDLLQ NTDGNSSSSI SLEQILRCLN SWSYEFPVEQ LLTVQPLINL VFETISNGNE 240
241 SDMEAFDSAI DCLCVILRES RDTTNEQLIS ALFHQLMLLQ EKLLPTLFTD HPLNDEYDDD 300
301 LLEGMTRLFV EAGEAWSVVI SKNPDFFKPM VLVLLMLTCK NEDLDVVSYT FPFWFNFKQS 360
361 LVLPRYQESR KAYSDIFVKL INGIITHLQY PSGQFSSKEE EDKFKDFRYH MGDVLKDCTA 420
421 VVGTSEALSQ PLIRIKSAIE NNNSWQIMEA PLFSLRTMAK EISLTENTIL PEIIKIICNL 480
481 PEQAKIRYAS TLVLGRYTEW TAKHPELLEV QLQYIFNGFQ LHEGSSDMQS IITASSHALM 540
541 FFCSDCSKLL VGYIDQLINF FLNVQSSIDI ESQFELCQGL SAVINNQPEA KVSVIFQKLV 600
601 DDNLRQIEAL IPQWKANPTL LAPQIADKID LLYALFEELK PRYNYPQQGS EPLLPRIEFI 660
661 WKALRTLLVD AGAMTDSIIV ERVAKLLRRI FERFHVFCEP ILPSVAEFLI QGYLTTGFGS 720
721 YLWCSGSLIV IFGDDESFPI SPSLKDAVWK FALSQCETFI LNFNKFDKLQ LNDYHEAIID 780
781 FFSLISDLIM FYPGAFLNST ELLGPVLNVA LECVNKLDNY DAYICILRCL DDIISWGFKT 840
841 PPISTVSIEI VPDEWRKQVI NEVVIAHGNQ LILVLFIGLV TTFENTAHSD AISCIVKCLR 900
901 ILTEANNNDA TICIDWIYKV VEQLGQVTLN ERDNLAKAVV EGLNSKDYRK VREGIRAFVG 960
961 WYSRKNINSR FE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
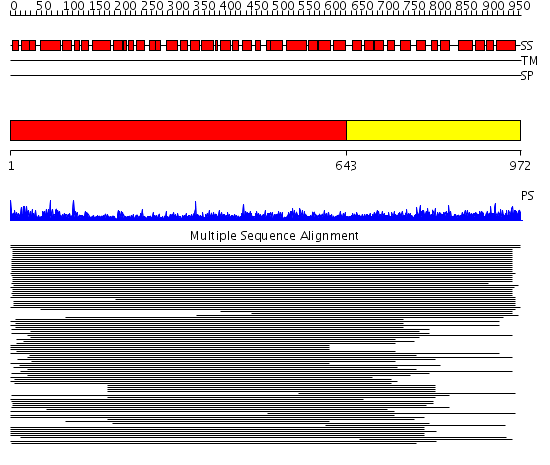
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..642] | 13.150996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [643..972] | 1.118943 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)