













| General Information: |
|
| Name(s) found: |
CYPR_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 318 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWLKSLLLCL YSLVLCQVHA APSSGKQITS KDVDLQKKYE PSPPATHRGI ITIEYFDPVS 60
61 KSMKEADLTF ELYGTVVPKT VNNFAMLAHG VKAVIEGKDP NDIHTYSYRK TKINKVYPNK 120
121 YIQGGVVAPD VGPFTVYGPK FDDENFYLKH DRPERLAMAY FGPDSNTSEF IITTKADGNE 180
181 ELDGKSVVFG QITSGLDQLM DAIQYTETDE YGKPQHELRF LYFVLEILKI SNILDLHAAY 240
241 TEKVEKFRNG DVSVGSTLEN IFRNDKAYTP LTTSTGTTAY DLNHPISRAL MCLTVLGLCF 300
301 IAYKGMHEKP HTVSLRHK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
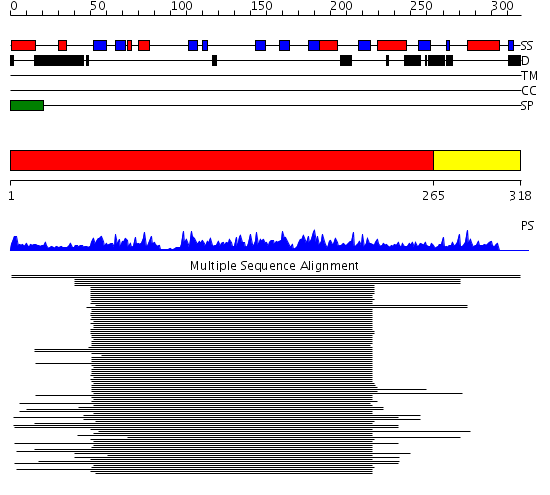
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..264] | 268.264245 | Cyclophilin (eukaryotic) | |
| 2 | View Details | [265..318] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|