













| General Information: |
|
| Name(s) found: |
UBP6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 482 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein deubiquitination
[TAS]
|
| Molecular Function: |
ubiquitin-specific protease activity
[IGI][ISS]
calmodulin binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTVSVKWQK KVLDGIEIDV SLPPYVFKAQ LYDLTGVPPE RQKIMVKGGL LKDDGDWAAI 60
61 GVKDGQKLMM MGTADEIVKA PEKAIVFAED LPEEALATNL GYSAGLVNLG NTCYMNSTVQ 120
121 CLKSVPELKS ALSNYSLAAR SNDVDQTSHM LTVATRELFG ELDRSVNAVS PSQFWMVLRK 180
181 KYPQFSQLQN GMHMQQDAEE CWTQLLYTLS QSLKAPTSSE GADAVKALFG VNLQSRLHCQ 240
241 ESGEESSETE SVYSLKCHIS HEVNHLHEGL KHGLKGELEK TSPALGRTAL YVKESLIDSL 300
301 PRYLTVQFVR FFWKRESNQK AKILRKVDYP LVLDIFDLCS EDLRKKLEAP RQKLREEEGK 360
361 KLGLQTSAKS GSKDSDVKMT DAEASANGSG ESSTVNPQEG TSSEKETHMT GIYDLVAVLT 420
421 HKGRSADSGH YVAWVKQESG KWIQYDDDNP SMQREEDITK LSGGGDWHMA YITMYKARFV 480
481 SM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
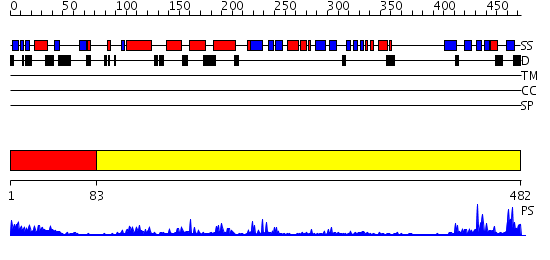
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 11.39794 | Solution Structure of the N-terminal Ubiquitin-like Domain of Mouse Ubiquitin Specific Protease 14 (USP14) | |
| 2 | View Details | [83..482] | 49.69897 | Structure of USP14, a proteasome-associated deubiquitinating enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)