













| General Information: |
|
| Name(s) found: |
gi|22329645
[NCBI NR]
gi|22136930 [NCBI NR] gi|18176246 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1373 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
pentose-phosphate shunt
[IEA]
glycolysis [IEA] metabolic process [IEA] valine metabolic process [IEA] |
| Molecular Function: |
zinc ion binding
[IEA]
coenzyme binding [IEA] phosphogluconate dehydrogenase (decarboxylating) activity [IEA] oxidoreductase activity [IEA] catalytic activity [IEA] 3-hydroxyisobutyrate dehydrogenase activity [IEA] binding [IEA] fructose-bisphosphate aldolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGVVGFVGL DSFSFELASS LLRSGFKVQA FEISTELVEK FIELGGHKCD SPADVGKAAA 60
61 AVVVVLSHPD QIQDVIFGDE GVMKGLQKDA VLLLSSTIST LQLQKLEKQL TEKREQIFVV 120
121 DAYVLKGMSE LLDGKLMIIA SGRSDSITRA QPYLTAMCQN LYTFEGEIGA GSKVKMVNEL 180
181 LEGIHLVAAV EAISLGSQAG VHPWILYDII SNAAGNSWIY KNHIPLLLKD DIEGRFLDVL 240
241 SQNLAIVEDK AKSLPFPVPL LAVARQQLIS GISQMQGDDT ATSLAKISEK VLGVGILEAA 300
301 NRELYKPEDL AKEITTQAKP VNRIGFIGLG AMGFGMAAHL LKSNFSVCGY DVYKPTLVRF 360
361 ENAGGLAANS PAEVTKDVDV LVIMVTNEVQ AEDVLYGHLG AVEAIPSGAT VVLASTVSPA 420
421 FVSQLERRLE NEGKDLKLVD APVSGGVKRA AMGELTIMAS GTDEALKSAG LVLSALSEKL 480
481 YVIKGGCGAG SGVKMVNQLL AGVHIASAAE AMAFGARLGL NTRKLFNVIS NSGGTSWMFE 540
541 NRVPHMLDND YTPYSALDIF VKDLGIVTRE GSSRKVPLHI STVAHQLFLA GSAAGWGRID 600
601 DAGVVKVYET LAGIKVEGRL PVLKKQDLLK SLPAEWPSDP TTDIHRLNMG NSKTLVVLDD 660
661 DPTGTQTVHD VEVLTEWSVE SISEQFRKKP ACFFILTNSR SLSPEKASEL IKDICSNLCA 720
721 ASKEVGNADY TIVLRGDSTL RGHFPQEADA AVSILGEMDA WIICPFFLQG GRYTIDDVHY 780
781 VADSDRLVPA GETEFAKDAS FGYKSSNLRE WVEEKTAGVI PANSVQSISI QLLRKGGPDA 840
841 VCEFLCSLKK GSTCIVNAAS ERDMAVFAAG MIQAELKGRS FLCRTAASFV SALIGIIPKD 900
901 PVLPKDFESN KESSGALIVV GSYVPKTTKQ VEELQSQHNQ NLRSIEISVE KVALKSSEVR 960
961 DEEIRRAVEM ADAFLRAGRE TLIMSSRELI TGKTSSESLD INSKVSSALV EVVSQISTRP1020
1021 RYILAKGGIT SSDTATKALK ARRALVIGQA LAGVPVWKLG PESRHPGVPY IVFPGNVGNS1080
1081 TALAEVVKSW SVVAGRSTKE LLLNAEKGGY AVGAFNVYNL EGIEAVVAAA EEENSPAILQ1140
1141 VHPGAFKQGG IPLVSCCISA AEQARVPISV HFDHGTTKHE LLEALELGLD SVMVDGSHLS1200
1201 FTENLSYTKS ITELARSKNI MVEAELGRLS GTEDGLTVED YEAKLTNVNQ AQEFMETGID1260
1261 ALAVCIGNVH GKYPKSGPNL KLDLLKELHA LSSKKGVFLV LHGASGLSEN LIKECIENGV1320
1321 RKFNVNTEVR TAYMEALSSG KKTDIVDVMS ATKAAMKAVI ADKIRLFGSA GKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
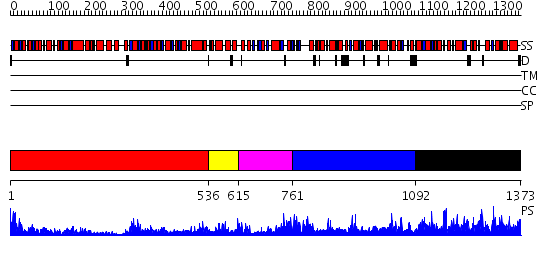
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..535] | 11.221849 | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form I (native2) | |
| 2 | View Details | [536..614] | 54.69897 | X-RAY CRYSTAL STRUCTURE OF TARTRONATE SEMIALDEHYDE REDUCTASE [SALMONELLA TYPHIMURIUM LT2] | |
| 3 | View Details | [615..760] | 3.06 | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE HYPOTHETICAL PROTEIN HI1011, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | |
| 4 | View Details | [761..1091] | 93.0 | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE HYPOTHETICAL PROTEIN HI1011, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | |
| 5 | View Details | [1092..1373] | 97.09691 | Class II fructose-1,6-bisphosphate aldolase from Thermus aquaticus in complex with cobalt |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)