













| General Information: |
|
| Name(s) found: |
CDPKT_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] plasma membrane [IDA] |
| Biological Process: |
protein amino acid phosphorylation
[IEA][ISS]
|
| Molecular Function: |
ATP binding
[IEA]
protein tyrosine kinase activity [IEA] calcium ion binding [IEA] protein kinase activity [IEA] calmodulin-dependent protein kinase activity [ISS] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGFCFSKFGK SQTHEIPISS SSDSSPPHHY QPLPKPTVSQ GQTSNPTSNP QPKPKPAPPP 60
61 PPSTSSGSQI GPILNRPMID LSALYDLHKE LGRGQFGITY KCTDKSNGRE YACKSISKRK 120
121 LIRRKDIEDV RREVMILQHL TGQPNIVEFR GAYEDKDNLH LVMELCSGGE LFDRIIKKGS 180
181 YSEKEAANIF RQIVNVVHVC HFMGVVHRDL KPENFLLVSN EEDSPIKATD FGLSVFIEEG 240
241 KVYRDIVGSA YYVAPEVLHR NYGKEIDVWS AGVMLYILLS GVPPFWGETE KTIFEAILEG 300
301 KLDLETSPWP TISESAKDLI RKMLIRDPKK RITAAEALEH PWMTDTKISD KPINSAVLVR 360
361 MKQFRAMNKL KKLALKVIAE NLSEEEIKGL KQTFKNMDTD ESGTITFDEL RNGLHRLGSK 420
421 LTESEIKQLM EAADVDKSGT IDYIEFVTAT MHRHRLEKEE NLIEAFKYFD KDRSGFITRD 480
481 ELKHSMTEYG MGDDATIDEV INDVDTDNDG RINYEEFVAM MRKGTTDSDP KLIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
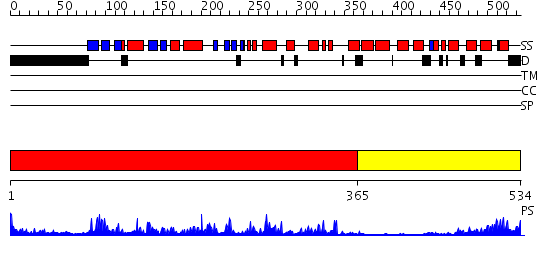
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..364] | 75.522879 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 2 | View Details | [365..534] | 26.39794 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)