













| General Information: |
|
| Name(s) found: |
PRS7B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 464 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
|
| Biological Process: |
protein catabolic process
[IEA]
|
| Molecular Function: |
ATPase activity
[ISS]
nucleotide binding [IEA] ATP binding [IEA] peptidyl-prolyl cis-trans isomerase activity [IEA] nucleoside-triphosphatase activity [IEA] hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIIRDIMEDP ENEGPHNMRS DFEPLLSVLR LRDYETGDII EFDSTEASEE ALSDITEFGS 60
61 TEASEAHFEE PYSARIKKVE KEINELAEKI CNLGIKESDT GLAPPNQWDL VSDKQMMQEE 120
121 QPLLVATCTQ IISPNTEDAK YVVDIKKIGK YVVGLGDKAS PTDIEAGMRV GVDQKKYQIQ 180
181 IPLPPKIDPS VTMMTVEEKP DATYSDIGGC KEQIEKIREV VELPMLHPEK FVRLGIDPPK 240
241 GVLCYGPPGS GKTLVARAVA NRTGACFIRV VGSELVQKYI GEGARMVREL FQMARSKKAC 300
301 ILFFDEIDAI GGARFDDGVG SDNEVQRTML EILYQLDGFD ARGNIKVLMA TNRPDILDPA 360
361 LLRPGRLDRK VEFCLPDLEG RTQIFKIHTR TMSCERDIRF ELLAGLCPNS TGADIRSVCI 420
421 EAGMYAIGAR RKSVTEKDFL DAVNKVVKGY QKFSATPKYM AYYI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
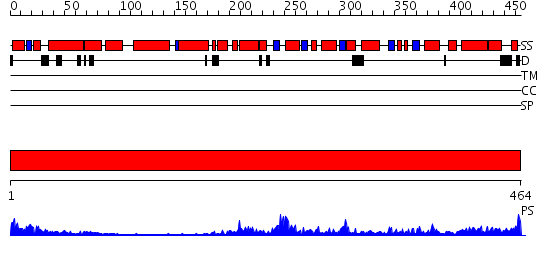
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..464] | 82.69897 | The crystal structure of murine p97/VCP at 3.6A |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)