













| General Information: |
|
| Name(s) found: |
MCM6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 831 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA replication initiation
[ISS]
DNA unwinding involved in replication [TAS] |
| Molecular Function: |
ATP binding
[IEA][ISS]
DNA binding [IEA][ISS] DNA-dependent ATPase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEAFGGFVMD EQAIQVENVF LEFLKSFRLD ANKPELYYEA EIEAIRGGES TMMYIDFSHV 60
61 MGFNDALQKA IADEYLRFEP YLRNACKRFV IEMNPSFISD DTPNKDINVS FYNLPFTKRL 120
121 RELTTAEIGK LVSVTGVVTR TSEVRPELLY GTFKCLDCGS VIKNVEQQFK YTQPTICVSP 180
181 TCLNRARWAL LRQESKFADW QRVRMQETSK EIPAGSLPRS LDVILRHEIV EQARAGDTVI 240
241 FTGTVVVIPD ISALAAPGER AECRRDSSQQ KSSTAGHEGV QGLKALGVRD LSYRLAFIAN 300
301 SVQIADGSRN TDMRNRQNDS NEDDQQQFTA EELDEIQQMR NTPDYFNKLV GSMAPTVFGH 360
361 QDIKRAVLLM LLGGVHKTTH EGINLRGDIN VCIVGDPSCA KSQFLKYTAG IVPRSVYTSG 420
421 KSSSAAGLTA TVAKEPETGE FCIEAGALML ADNGICCIDE FDKMDIKDQV AIHEAMEQQT 480
481 ISITKAGIQA TLNARTSILA AANPVGGRYD KSKPLKYNVN LPPAILSRFD LVYVMIDDPD 540
541 EVTDYHIAHH IVRVHQKHEA ALSPEFTTVQ LKRYIAYAKT LKPKLSPEAR KLLVESYVAL 600
601 RRGDTTPGTR VAYRMTVRQL EALIRLSEAI ARSHLEILVK PSHVLLAVRL LKTSVISVES 660
661 GDIDLSEYQD ANGDNMDDTD DIENPVDGEE DQQNGAAEPA SATADNGAAA QKLVISEEEY 720
721 DRITQALVIR LRQHEETVNK DSSELPGIRQ KELIRWFIDQ QNEKKKYSSQ EQVKLDIKKL 780
781 RAIIESLVCK EGHLIVLANE QEEAAEAEET KKKSSQRDER ILAVAPNYVI E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
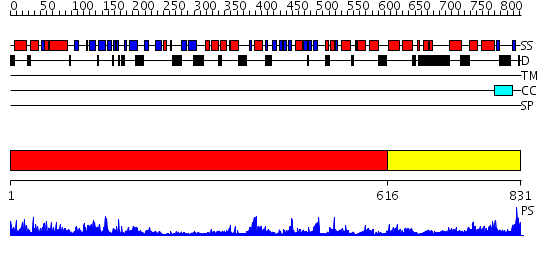
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..615] | 68.09691 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [616..831] | 62.39794 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)