













| General Information: |
|
| Name(s) found: |
gi|79327143
[NCBI NR]
gi|30681498 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMEKPPLAS GLARTRSEQL YETVAADIRS PHGSMDANGV PATAPAAVGG GGTLSRKSSR 60
61 RLMGMSPGRS SGAGTHIRKS RSAQLKLELE EVSSGAALSR ASSASLGLSF SFTGFAMPPE 120
121 EISDSKPFSD DEMIPEDIEA GKKKPKFQAE PTLPIFLKFR DVTYKVVIKK LTSSVEKEIL 180
181 TGISGSVNPG EVLALMGPSG SGKTTLLSLL AGRISQSSTG GSVTYNDKPY SKYLKSKIGF 240
241 VTQDDVLFPH LTVKETLTYA ARLRLPKTLT REQKKQRALD VIQELGLERC QDTMIGGAFV 300
301 RGVSGGERKR VSIGNEIIIN PSLLLLDEPT SGLDSTTALR TILMLHDIAE AGKTVITTIH 360
361 QPSSRLFHRF DKLILLGRGS LLYFGKSSEA LDYFSSIGCS PLIAMNPAEF LLDLANGNIN 420
421 DISVPSELDD RVQVGNSGRE TQTGKPSPAA VHEYLVEAYE TRVAEQEKKK LLDPVPLDEE 480
481 AKAKSTRLKR QWGTCWWEQY CILFCRGLKE RRHEYFSWLR VTQVLSTAVI LGLLWWQSDI 540
541 RTPMGLQDQA GLLFFIAVFW GFFPVFTAIF AFPQERAMLN KERAADMYRL SAYFLARTTS 600
601 DLPLDFILPS LFLLVVYFMT GLRISPYPFF LSMLTVFLCI IAAQGLGLAI GAILMDLKKA 660
661 TTLASVTVMT FMLAGGFFVK ASPLFLDFLC F |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
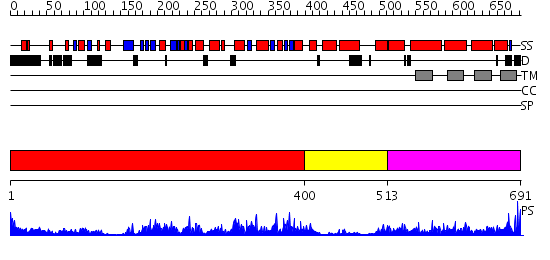
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..399] | 59.0 | No description for 2hydA was found. | |
| 2 | View Details | [400..512] | 72.0 | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius | |
| 3 | View Details | [513..691] | 19.107905 | No description for PF01061.15 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|