













| General Information: |
|
| Name(s) found: |
gi|59958334
[NCBI NR]
gi|30679610 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 628 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
microtubule-based movement
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [IEA][ISS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDSKTPAKT KLLDSSSISN VRVVLRVRPF LPREISDESC DGRSCVSVIG GDGGDTSEVA 60
61 VYLKDPDSCR NESYQLDAFY GREDDNVKHI FDREVSPLIP GIFHGFNATV LAYGATGSGK 120
121 TFTMQGIDEL PGLMPLTMST ILSMCEKTRS RAEISYYEVY MDRCWDLLEV KDNEIAVWDD 180
181 KDGQVHLKGL SSVPVKSMSE FQEAYLCGVQ RRKVAHTGLN DVSSRSHGVL VISVTSQGLV 240
241 TGKINLIDLA GNEDNRRTGN EGIRLQESAK INQSLFALSN VVYALNNNLP RVPYRETKLT 300
301 RILQDSLGGT SRALMVACLN PGEYQESLRT VSLAARSRHI SNNVSLNPKV ETPKVKIDME 360
361 AKLQAWLESK GKMKSAHRMM AIRSPLMGTN QTSISQSSVK KLLCHRSAIA ESAKLAGTGQ 420
421 RDAFVTARNL FGVETLAASH LWEPIRNLQL ASPTKEDERD TSGEENLLVS EASLRDNTLD 480
481 VEKKYTELSP LREALSPIDS NAKPNSAHGS SPFLKPMTPK TPFLSTNPEI MQMEGTCQKF 540
541 NAWSNNLKTS LIKEYIHFLN TANREELREL KGIGQKMAEY IIELRETSPL KSLTDLEKIG 600
601 FTSRQVHNLF KKATEGVLEK PVSASTTP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
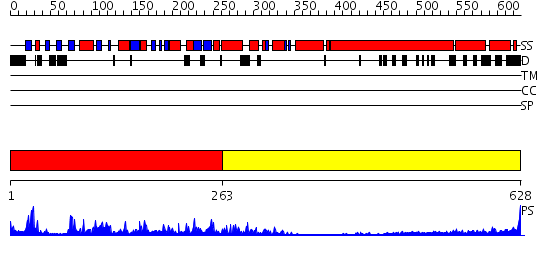
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..262] | 85.69897 | Kinesin | |
| 2 | View Details | [263..628] | 7.0 | No description for 2dfsA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)