













| General Information: |
|
| Name(s) found: |
DMA1 /
YHR115C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 416 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IGI establishment of mitotic spindle orientation [IGI septin ring assembly [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTNTVPSSP PNQTPPAASG IATSHDHTKF NNPIRLPISI SLTINDTPNN NSNNNSVSNG 60
61 LGILPSRTAT SLVVANNGSA NGNVGATAAA AATVETNTAP AVNTTKSIRH FIYPPNQVNQ 120
121 TEFSLDIHLP PNTSLPERID QSTLKRRMDK HGLFSIRLTP FIDTSSTSVA NQGLFFDPII 180
181 RTAGAGSQII IGRYTERVRE AISKIPDQYH PVVFKSKVIS RTHGCFKVDD QGNWFLKDVK 240
241 SSSGTFLNHQ RLSSASTTSK DYLLHDGDII QLGMDFRGGT EEIYRCVKMK IELNKSWKLK 300
301 ANAFNKEALS RIKNLQKLTT GLEQEDCSIC LNKIKPCQAI FISPCAHSWH FHCVRRLVIM 360
361 NYPQFMCPNC RTNCDLETTL ESESESEFEN EDEDEPDIEM DIDMEINNNL GVRLVD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
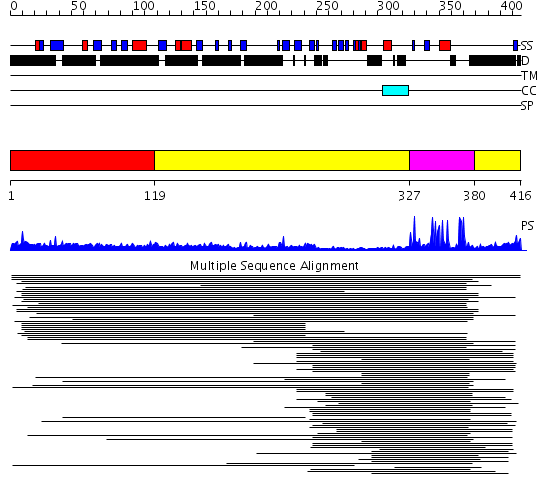
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [119..326] [380..416] |
6.0 | Cbl; N-terminal domain of cbl (N-cbl); CBL | |
| 3 | View Details | [327..379] | 6.0 | Cbl; N-terminal domain of cbl (N-cbl); CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)