













| General Information: |
|
| Name(s) found: |
SLX5 /
YDL013W
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 619 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein sumoylation
[IGI telomere maintenance [IMP response to DNA damage stimulus [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSDTNGRTK SNNSPSDNNP NETVILIDSD KEEDASIREA NLPVRLYPDR RVGRRRDALN 60
61 RFVRSDSRSR NSQRTHITAS SERPDFQANN DDITIIREVG RFFGDDGPID PSAHYVDLDQ 120
121 EPGSETLETP RTIQVDNTNG YLNDNGNNNE SDDGLTIVEE RTTRPRVTLN LPGGERLEVT 180
181 ATTTDIPIRR SFEFQEDLGA SRRQLLRRSA TRARNLFVDR SDENDEDWTD DTHNLPEAIQ 240
241 RARRESRMRM SRRIAERQRR VQQQRVSSDE NISTSIRLQS IRERIQSYTP DIRSAFHRAE 300
301 SLHEFRSILQ NVAPITLQEC EEELMALFTE FRNQLLQNWA IDRVRNTQEE ALRLHREALE 360
361 RQERTAGRVF HRGTLRESIT NYLNFNGEDG FLSRLWSGPA LSDADEERHT QNIIDMIQER 420
421 EERERDVVMK NLMNKTRAQQ EEFEARAASL PEGYSASFDT TPKMKLDITK NGKEETIIVT 480
481 DDDLAKTLED IPVCCLCGAE LGVGIPDDFT GISQKDRGVS FEGLVSKYKF HCPYQTLARP 540
541 SMLDRDLSKR TFIASCGHAF CGRCFARIDN AKKKSKMPKK KLAQLKGSAH PDNYGPKLCP 600
601 ADSCKKLIRS RGRLKEVYF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
GSG1 POL4 RPN1 RPN10 RPN11 RPN12 RPN13 RPN2 RPN5 RPN6 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 SEM1 SLX5 SPG5 SWA2 UBP6 |
| View Details | Krogan NJ, et al. (2006) |

|
ABF2 ARG82 CKI1 CMR1 HHF1, HHF2 HTA1 HTA2 HTB1 HTB2 NUP188 PPH3 PSY2 PSY4 RRD1 SLX5 SLX8 SPT5 STP1 TOP2 |
| View Details | Ho Y, et al. (2002) |
|
PTP2 SLX5 SSK1 SSK2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
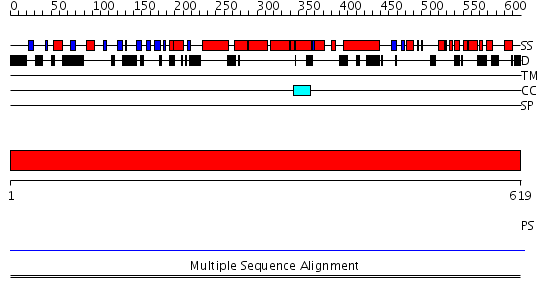
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..619] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [129..224] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [225..400] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [401..619] | 1.21 | Effector domain of rabphilin-3a |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)