













| General Information: |
|
| Name(s) found: |
SKI7 /
YOR076C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA exosome (RNase complex) [IDA |
| Biological Process: |
response to exogenous dsRNA
[IMP mRNA catabolic process [IDA |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLLEQLARK RIEKSKGLLS ADQSHSTSKS ASLLERLHKN RETKDNNAET KRKDLKTLLA 60
61 KDKVKRSDFT PNQHSVSLSL KLSALKKSNS DLEKQGKSVT LDSKENELPT KRKSPDDKLN 120
121 LEESWKAIKE MNHYCFLKND PCINQTDDFA FTNFIIKDKK NSLSTSIPLS SQNSSFLSLK 180
181 KHNNELLGIF VPCNLPKTTR KVAIENFNRP SPDDIIQSAQ LNAFNEKLEN LNIKSVPKAE 240
241 KKEPINLQTP PTESIDIHSF IATHPLNLTC LFLGDTNAGK STLLGHLLYD LNEISMSSMR 300
301 ELQKKSSNLD PSSSNSFKVI LDNTKTEREN GFSMFKKVIQ VENDLLPPSS TLTLIDTPGS 360
361 IKYFNKETLN SILTFDPEVY VLVIDCNYDS WEKSLDGPNN QIYEILKVIS YLNKNSACKK 420
421 HLIILLNKAD LISWDKHRLE MIQSELNYVL KENFQWTDAE FQFIPCSGLL GSNLNKTENI 480
481 TKSKYKSEFD SINYVPEWYE GPTFFSQLYL LVEHNMNKIE TTLEEPFVGT ILQSSVLQPI 540
541 AEINYVSLKV LINSGYIQSG QTIEIHTQYE DFHYYGIVSR MKNSKQILET NTKNNISVGL 600
601 NPDILEVLVK IHNTEDFTKK QFHIRKGDII IHSRKTNTLS PNLPNTLKLL ALRLIKLSIQ 660
661 THALSDPVDL GSELLLYHNL THNAVKLVKI LGTNDISINP NQSLIVEVEI IEPDFALNVI 720
721 DSKYITNNIV LTSIDHKVIA VGRIACQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CSL4 DIS3 LRP1 MPP6 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI3 SKI6 SKI7 SRP1 |
| View Details | Krogan NJ, et al. (2006) |
|
CSL4 DDI1 DIS3 ENT3 LRP1 MPP6 MTR3 OSM1 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 YBL055C |
| View Details | Gavin AC, et al. (2006) |
|
CSL4 DIS3 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SKI6 SKI7 |
| View Details | Gavin AC, et al. (2002) |

|
CMD1 CSL4 CTR9 DIS3 ECM16 GCN1 LRP1 MAM33 MLC1 MTR3 MYO2 NMD5 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SAM1 SHE4 SKI2 SKI3 SKI6 SKI7 SRP1 UTP10 UTP20 UTP22 YIR035C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
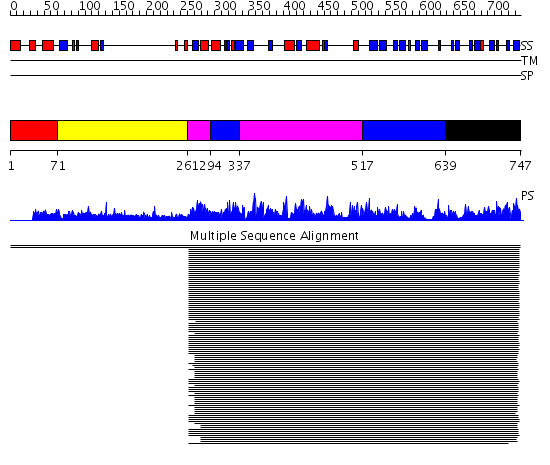
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..260] | 5.516997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [261..293] [337..516] |
1110.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain | |
| 4 | View Details | [294..336] [517..638] |
1110.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain | |
| 5 | View Details | [639..747] | 1110.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)