













| General Information: |
|
| Name(s) found: |
SQS1 /
YNL224C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRHSHYQG SRRRHARGSN SKKAGRGNAK GIQGRKIKKK PTPTNSWHNS SIPLGEGDLD 60
61 DVGADFNPGR AFISPKTIED YYFGRDAKSR SMKMGGLRPG NRYDSSTDLQ AGRAAFRKRP 120
121 MQFVKAKEVY DPSHNMIQKL RAKNETKNSE EIVEREADVF EEPGKMTSDV EYINNEDSEN 180
181 EDDDSQNSPS TDHSLSSNES KVEDGDLFFV DEEAQQSPDL TKIKRVCIEE IARPREVAIE 240
241 FDPILTIGKV ELSVSEGNES KEISVDVPNK GNKTYHPFAG YISNVLHGMH TSDSDNDELD 300
301 YEIETENNSE PLYESSASSE VDQGFNYVGQ RHNSRADNNL LPSPSPQLTE DIKCLSINGT 360
361 KTFEGNNDNL PSPASEELEF GFKEEDFVIN TNDIVVSNIR MGGVDNSYYL RCYRLLGDYD 420
421 FHWIDQDLLT DFVVDELGLP EDRLPAYLNF IKNSLIPKIE PAEPTYSDIP ISDSSDEGDS 480
481 YEGDSYEDDE DMASSVVHSD IEEGLDDLIA YTLKHDTERF KTFETKSLET KGKGKKKKLL 540
541 IDDALALDTE TLETLQSKFS KRIETKAKKR KAKEDFIDQE NRNSNDMLKK YPYGLHIQNI 600
601 KDEFESFLSR NNDRLTFPPL DPHGNKTVMK IAKHYNMKSS KIGKANHTSV VVEKIKKTKW 660
661 SSPNYSLIDQ LMRQRPVFMR IDIRRPREEQ AAFERTKTIR GKFHVKEGEI VGQNAPEIGN 720
721 ENIGRRMLEK LGWKSGEGLG IQGNKGISEP IFAKIKKNRS GLRHSES |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CWC23 PRP43 SQS1 |
| View Details | Krogan NJ, et al. (2006) |

|
CWC23 PRP43 SPP382 SQS1 |
| View Details | Gavin AC, et al. (2006) |

|
PRP43 SQS1 |
| View Details | Gavin AC, et al. (2002) |

|
ASC1 CDC33 CLF1 CWC23 DCP2 DHH1 DIB1 ERB1 KRS1 LSM1 LSM2 LSM3 LSM4 LSM5 LSM6 LSM7 NAM7 NOG2 NOP1 NUG1 PAT1 PRP24 PRP31 PRP38 PRP4 PRP43 PRP46 PRP6 SMB1 SMD2 SNU114 SNU23 SNU66 SPP382 SQS1 SRO9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
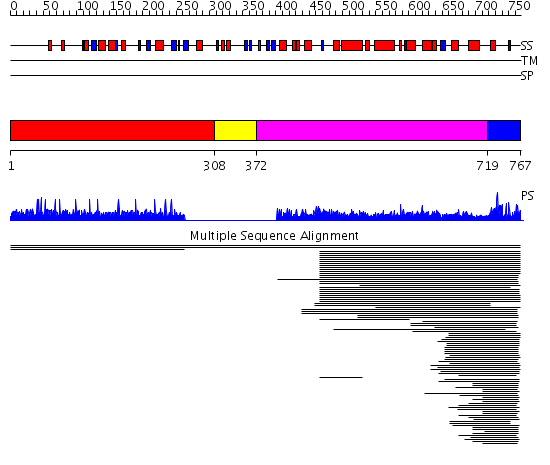
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..307] | 1.002856 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [308..371] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [372..718] | 1.075997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [719..767] | 16.376751 | G-patch domain No confident structure predictions are available. | |
| 5 | View Details | [531..683] | 1.38 | SmuBP-2 | |
| 6 | View Details | [684..767] | 50.185997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)