













| General Information: |
|
| Name(s) found: |
YKT6 /
YKL196C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 200 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA mitochondrion [IDA membrane [IDA fungal-type vacuole [IDA |
| Biological Process: |
vesicle fusion
[TAS intra-Golgi vesicle-mediated transport [TAS |
| Molecular Function: |
receptor signaling protein serine/threonine kinase activity
[TAS palmitoyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRIYYIGVFR SGGEKALELS EVKDLSQFGF FERSSVGQFM TFFAETVASR TGAGQRQSIE 60
61 EGNYIGHVYA RSEGICGVLI TDKEYPVRPA YTLLNKILDE YLVAHPKEEW ADVTETNDAL 120
121 KMKQLDTYIS KYQDPSQADA IMKVQQELDE TKIVLHKTIE NVLQRGEKLD NLVDKSESLT 180
181 ASSKMFYKQA KKSNSCCIIM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
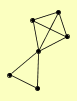
|
DSL1 SEC17 SEC39 TIP20 VTI1 YKT6 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
NYV1 YKT6 |
| View Details | Gavin AC, et al. (2006) |
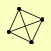
|
PEP12 SEC17 VTI1 YKT6 |
| View Details | Gavin AC, et al. (2002) |
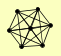
|
SEC17 SLY1 TLG1 TLG2 VPS45 VTI1 YKT6 |
| View Details | Qiu et al. (2008) |
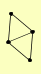
|
NYV1 SEC22 VAM7 YKT6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample: hx1 | Hao Xu, et al. (2010) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
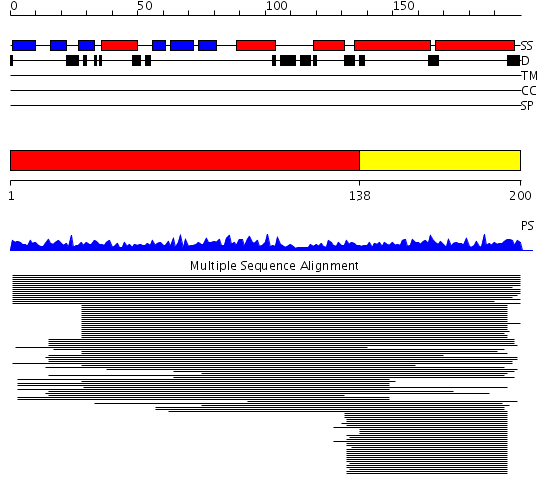
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 798.68867 | Synaptobrevin homolog 1 ykt6 | |
| 2 | View Details | [138..200] | 48.9897 | Endobrevin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)