













| General Information: |
|
| Name(s) found: |
VAM7 /
YGL212W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 316 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA |
| Biological Process: |
vesicle fusion
[TAS telomere maintenance [IMP Golgi to vacuole transport [TAS |
| Molecular Function: |
receptor signaling protein serine/threonine kinase activity
[TAS phosphatidylinositol-3-phosphate binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAANSVGKMS EKLRIKVDDV KINPKYVLYG VSTPNKRLYK RYSEFWKLKT RLERDVGSTI 60
61 PYDFPEKPGV LDRRWQRRYD DPEMIDERRI GLERFLNELY NDRFDSRWRD TKIAQDFLQL 120
121 SKPNVSQEKS QQHLETADEV GWDEMIRDIK LDLDKESDGT PSVRGALRAR TKLHKLRERL 180
181 EQDVQKKSLP STEVTRRAAL LRSLLKECDD IGTANIAQDR GRLLGVATSD NSSTTEVQGR 240
241 TNNDLQQGQM QMVRDQEQEL VALHRIIQAQ RGLALEMNEE LQTQNELLTA LEDDVDNTGR 300
301 RLQIANKKAR HFNNSA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
VAM3 VAM7 |
| View Details | Qiu et al. (2008) |
|
ATG21 NYV1 VAM7 VMA9 VTI1 YKT6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample: hx1 | Hao Xu, et al. (2010) | |
| View Run | sample: hx2 | Hao Xu, et al. (2010) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
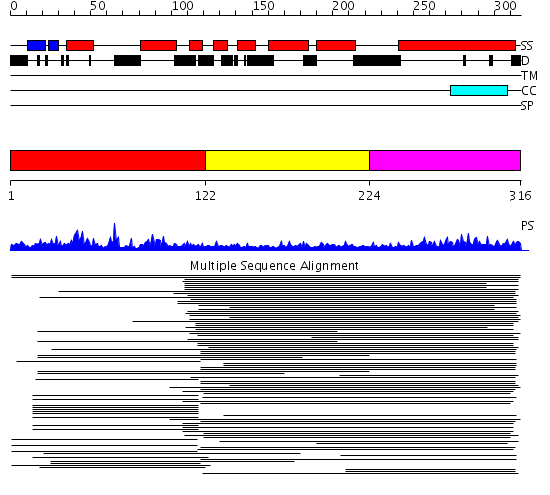
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 616.9897 | Vam7p | |
| 2 | View Details | [122..223] | 6.0 | Synaptosomal-assosiated protein SNAP-25 | |
| 3 | View Details | [224..316] | 11.045757 | Synaptosomal-assosiated protein SNAP-25 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)