













| General Information: |
|
| Name(s) found: |
PIB2 /
YGL023C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 635 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA late endosome [TAS |
| Biological Process: |
vesicle-mediated transport
[ISS |
| Molecular Function: |
phosphatidylinositol binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTALHSVSKT PAIKEEEEDG DERDGRGVPL GPRNHDYRGR KGDEESGADT VTSPITFEKK 60
61 KIAPRASTHS EQSILSSISL KSMVNQHRQQ QLQQESSTGA GTGFVDRKQQ IQSPAMVSIL 120
121 RKNSAEENVR SSHSSKLGEG QIDGRKASAS KEIGKTLPFT DDQRSNPELD PTNSVVDVSR 180
181 GKNTKSKTVF NELEDDADDD DEVRQKNLTT QALRKLSSFK MNASSNLRLS KENKAKESSS 240
241 SSTSSVSSSS TSKVENIVDK LTTTNSSSMS QLRFGNTNVI IDSVNHAAKP PHQQMLRKPS 300
301 LEFLPQPASS TNLNFNSNKH KSNVRQISNP KKPLYIPAVL RKVSETNITN DDLLNATLSS 360
361 YYKKASNLEH GFNPSKSQSA SVQNANNLRI ISSQSSVQSN TSSILESYKN KISSYLFPNS 420
421 IPNSDRINLI PTISNRNSAR VNPPTKDHWI PDSKRNSCRY CHKPFTLWER KHHCRHCGDI 480
481 FCQDHLRHWL YLDSQANFIM INELNNGGIN GGGTLCKICD DCLVEYENLS TTNHNANTNE 540
541 DNINVEEGED DDNDNRKKLR NYYKNRQMNA LFRPKKGGSS QEHATVDRDT TTPIQVKSND 600
601 EEADNENTGG EQEEGNDVLG SVIGSVPANW NWSSF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
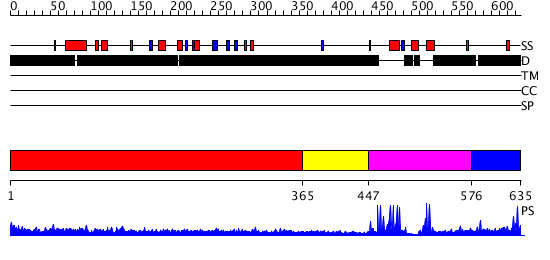
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..364] | 13.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [365..446] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [447..575] | 93.751666 | vps27p protein | |
| 4 | View Details | [576..635] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)