













| General Information: |
|
| Name(s) found: |
SKN7 /
YHR206W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 622 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
response to oxidative stress
[TAS transcription [IDA regulation of cell size [IMP response to osmotic stress [TAS response to singlet oxygen [IMP |
| Molecular Function: |
transcription factor activity
[IDA two-component response regulator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFSTINSNV NKTTGDSNNN TTENSSTADL LGMDLLQSGP RLMNTMQPNN SSDMLHINNK 60
61 TNNVQQPAGN TNISSANAGA KAPANEFVRK LFRILENNEY PDIVTWTENG KSFVVLDTGK 120
121 FTTHILPNHF KHSNFASFVR QLNKYDFHKV KRSPEERQRC KYGEQSWEFQ HPEFRVHYGK 180
181 GLDNIKRKIP AQRKVLLDES QKALLHFNSE GTNPNNPSGS LLNESTTELL LSNTVSKDAF 240
241 GNLRRRVDKL QKELDMSKME SYATKVELQK LNSKYNTVIE SLITFKTINE NLLNNFNTLC 300
301 STLANNGIEV PIFGDNGNRN PTGNTNPATT TAIQSNNNTN NASPATSTVS LQLPNLPDQN 360
361 SLTPNAQNNT VTLRKGFHVL LVEDDAVSIQ LCSKFLRKYG CTVQVVSDGL SAISTLEKYR 420
421 YDLVLMDIVM PNLDGATATS IVRSFDNETP IIAMTGNIMN QDLITYLQHG MNDILAKPFT 480
481 RDDLHSILIR YLKDRIPLCE QQLPPRNSSP QTHSNTNTAN SNPNTINEQS LAMLPQDNPS 540
541 TTTPVTPGAS ISSAQHVQQG QQEQQHQIFH AQQQQQHHNA IANARSDVAI PNLEHEINTV 600
601 PHSSMGSTPQ LPQSTLQENQ LS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
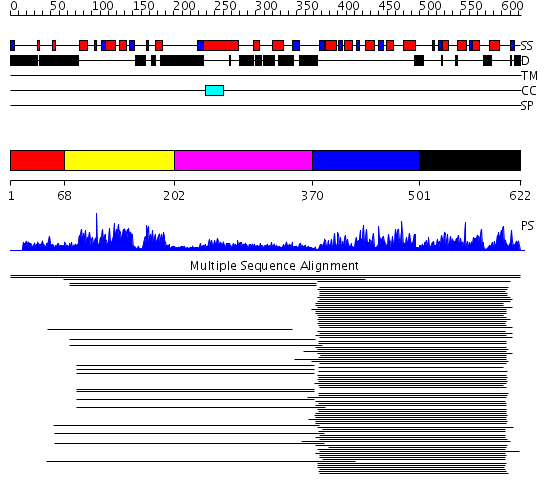
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [68..201] | 200.9897 | Heat-shock transcription factor | |
| 3 | View Details | [202..369] | 25.748997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [370..500] | 122.232149 | PhoB; PhoB receiver domain | |
| 5 | View Details | [501..622] | 122.232149 | PhoB; PhoB receiver domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)