













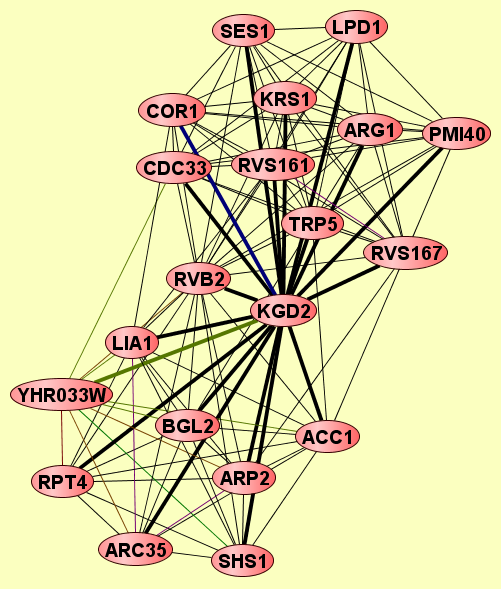
| Protein: | KGD2 |
| From Publication: | Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002 Jan 10;415(6868):180-3. |
| Complexes containing KGD2: | 2 |
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 12 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Rvs167. The topolgies of protein complexes in this experiment are unknown. | ARG1 CDC33 COR1 KGD2 KRS1 LPD1 PMI40 RVB2 RVS161 RVS167 SES1 TRP5 |
| View GO Analysis | 10 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Shs1. The topolgies of protein complexes in this experiment are unknown. | ACC1 ARC35 ARP2 BGL2 KGD2 LIA1 RPT4 RVB2 SHS1 YHR033W |