













| General Information: |
|
| Name(s) found: |
Psi-PC /
FBpp0086222
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 797 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U1 snRNP
[IPI]
|
| Biological Process: |
regulation of alternative nuclear mRNA splicing, via spliceosome
[IMP]
negative regulation of nuclear mRNA splicing, via spliceosome [IDA][IMP] spermatogenesis [IMP] regulation of nuclear mRNA splicing, via spliceosome [IMP] mRNA processing [IDA] |
| Molecular Function: |
mRNA binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDFQQQGIN QSQALAQAIQ RAKQIAAKIQ PSQQGGGTAG PSPPSSGGGP GFKRINDDGD 60
61 SGPESKRSLG SPEYANNSSN MSSGSGGGGG GGGGPGGASI TQAIAQAAAV AARLAASAGT 120
121 SCEEQIRLPE SVAGAFMGRS SNDTITHIQA ESGVKVQVMQ DQDRVIMLRG QRDTVTKGRE 180
181 MIQNMANRAG GGQVEVLLTI NMPPPGPSGY PPYQEIMIPG AKVGLVIGKG GDTIKQLQEK 240
241 TGAKMIIIQD GPNQELIKPL RISGEAQKIE HAKQMVLDLI AQKDAQGQQQ GGRGGGGGGG 300
301 GPGMGFNNFN NGNGGESTEV FVPKIAVGVV IGKGGDMIRK IQTECGCKLQ FIQGKNDEMG 360
361 DRRCVIQGTR QQVDDAKRTI DGLIENVMQR NGMNRNGNGG GGGPGGDSGN SNYGYGYGVN 420
421 HAQGGREEIT FLVPASKCGI VIGRGGETIK LINQQSGAHT EMDRNASNPP NEKLFKSKGT 480
481 TDQVEAARQM ISEKINMELN VISRKPIGGG PGGGGGNSGG GQNNSHHNQQ QGGYGGQNQM 540
541 QGGDPNSGAG YQQQPWGGPY GQQGWDPSGQ QQQMAMANQG GSAAGGASGG QDYSAQWIEY 600
601 YKQMGCHREA EMIEQQMKAK QAGGSSGPVQ QQQPQQQQQS QQQTGGAGAG GADYSAQWAE 660
661 YYRSVGKIEE AEAIEKTLKN KQQNGSGGQS STPNPSQGGS GQQQPNPAAA AAAAAAAAAA 720
721 AGGYGQSMTP TQYAQYSQYY AAAAAAGGQP QGAPQPGGGQ SGGPPGNYPG NYPGAGYGGY 780
781 PGAPGQQQQK SHKNDNH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
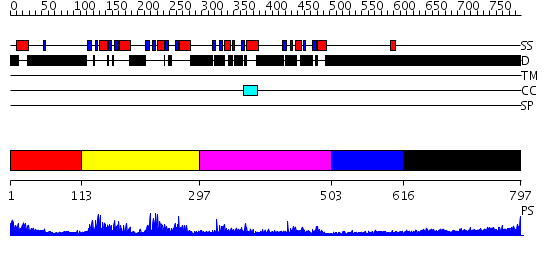
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [113..296] | 27.522879 | No description for 2annA was found. | |
| 3 | View Details | [297..502] | 19.0 | Far upstream binding element, FBP, KH3 and KH4 domains | |
| 4 | View Details | [503..615] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [616..797] | 7.0 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)