













| General Information: |
|
| Name(s) found: |
heph-PG /
FBpp0085246
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 615 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
ribonucleoprotein complex [IDA] cytoplasm [IDA] |
| Biological Process: |
negative regulation of oskar mRNA translation
[IMP]
Notch signaling pathway [IMP] spermatid development [IMP] imaginal disc-derived wing margin morphogenesis [IMP] oocyte microtubule cytoskeleton polarization [IMP] imaginal disc-derived wing vein morphogenesis [IMP] nuclear mRNA splicing, via spliceosome [ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
poly-pyrimidine tract binding [ISS][NAS] translation repressor activity, nucleic acid binding [IMP] mRNA binding [ISS] mRNA 3'-UTR binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MREFVATMMS CPIPMPMPIS SLHKSEFEPV NIGVKRGSDE LLSQAAVMAP ASDNNNQDLA 60
61 TKKAKLEPGT VLAGGIAKAS KVIHLRNIPN ESGEADVIAL GIPFGRVTNV LVLKGKNQAF 120
121 IEMADEISAT SMVSCYTVTP PQMRGRMVYV QFSNHRELKT DQGHNNSTAH SDYSVQSPAS 180
181 GSPLPLSAAA NATSNNANSS SDSNSAMGIL QNTSAVNAGG NTNAAGGPNT VLRVIVESLM 240
241 YPVSLDILHQ IFQRYGKVLK IVTFTKNNSF QALIQYPDAN SAQHAKSLLD GQNIYNGCCT 300
301 LRIDNSKLTA LNVKYNNDKS RDFTNPALPP GEPGVDIMPT AGGLMNTNDL LLIAARQRPS 360
361 LSGDKIVNGL GAPGVLPPFA LGLGTPLTGG YNNALPNLAA FSLANSGALQ TTAPAMRGYS 420
421 NVLLVSNLNE EMVTPDALFT LFGVYGDVQR VKILYNKKDS ALIQMAEPQQ AYLAMSHLDK 480
481 LRLWGKPIRV MASKHQAVQL PKEGQPDAGL TRDYSQNPLH RFKKPGSKNY QNIYPPSATL 540
541 HLSNIPSSCS EDDIKEAFTS NSFEVKAFKF FPKDRKMALL QLLSVEEAVL ALIKMHNHQL 600
601 SESNHLRVSF SKSNI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
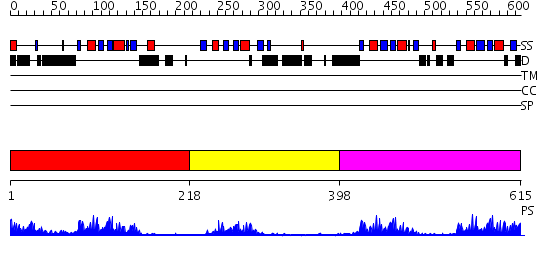
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 16.154902 | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | |
| 2 | View Details | [218..397] | 12.221849 | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | |
| 3 | View Details | [398..615] | 44.0 | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)