













| General Information: |
|
| Name(s) found: |
FBpp0306396
[FlyBase]
FBpp0306393 [FlyBase] e(y)3-PA / FBpp0074531 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2006 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome
[IDA]
heterochromatin [IDA] nucleus [IDA] euchromatin [IDA] |
| Biological Process: |
gene silencing
[IMP]
negative regulation of transcription, DNA-dependent [IMP] positive regulation of transcription, DNA-dependent [IMP] oogenesis [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] transcription regulator activity [IMP] chromatin binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDLRQQQMV AATSSSGSES GTAVESAAAT STAGSAGAAG RPQSNCSANS NAKSVAASST 60
61 SEEEQRVSST SSPAQRDQQL NADRDREQEP EPQQQQQREE ALQHQHNQPG HITSTTASPP 120
121 PTLPPPTTPC DDAPSTTGAS ASASSASGEA PSAASAAGAA GGPMAATALE VESEERDGHK 180
181 IILKLSKHAN PNSNANESQP GGDERRVEPL RIQLPCGGAE GGLVAKQQDA FDADASSCSS 240
241 SCAEDEVATT LGQQLRNTPH IVPKLTIRAA NERRVGSVVP KLTIKLPENP AASGSNSNSG 300
301 SCSAAVSGAQ SAMPAKNDAH LSSLSPASAS SSSASSSSSS SSSSSLAEMQ TVPKLMIKTT 360
361 LAGSSCISSS EELPQQQQQI PKLTIKTGGG GQEHVHTVIM THDLNNAQSI PKLTIKTKSI 420
421 EMIEDEQAAK LEQQLQPLPK LTIKNLCSPK HKVRAVLEEK PPTAASKLAI PKPTPNPTPA 480
481 PMTNGGESNS SSQEFCGFSD PDADIAAPAS DDVRRNSDDM VIDDSLSKEH DPKIFHNLPP 540
541 MPASNGIASG GSKASKASKS AQSQHNVVDM VDLTSSPSPG SSPTHVPNNF TGRISPKGLL 600
601 IDCLRMQTAI GCYVPEEADS RVRPAASPNS NILLSQLTAP AKSFTTTTPK SKSSKYPQLT 660
661 ERLMANGSGT GGGSVIAAES EAVVGPLPAA NPAQSIIESI EILDTPDGSP RVAYMDEDSN 720
721 PMLNKHLQTI HKLAMDHGVE QRMDTQDNNN ENHLKRTNSE GNESPSSRLP PSKQRRLDND 780
781 ENDQQTQNCH DPARKCSESL QALPPSHRSR KQKHERILNT DLEGSLFPPT QQQSISTPDQ 840
841 NGALSSEVEG EDAKAQDPLE PATPQPPPVA TPAGTGKKRG RPKRIQNQSS PDGGGAVTPK 900
901 PGTPQEEANS AVKSRRVQLL RKRLAIDMVS VGQEQADMKA KEKESSVGVP NARVEDGLAE 960
961 TLESPKTRDH RPLRATRRTT TSTNTNLQPT PKSTRKRQSK ASVQQSQLPP PTMAQFGSSE1020
1021 SESNNNNNSS ISFALSAQID LTMCSSSSST SSGAAANQQV IGGSGSSSML PPTTILSSSD1080
1081 PLPDVIFQPN DFSSIMATQQ LRSSRPSSIS CGSTGGSQPD CHDEDNYTSA LDNSGDETAL1140
1141 PMPPTRGRGR GRRSRGGRGR GSSSVDRAVS VGGTASTTPA TQPRAPRMSR GASAVAKAIA1200
1201 MSRPRCVGGL KHQPDPERLK GLFSPSPQVF EEDTRMSADL SNSNQSMASL MEPPLTPQKQ1260
1261 PDFLNNEESQ SSMVSNVSMM DSNQGQSADA ILGSALKRPK KKKMETCVAE DTDYSASSIA1320
1321 EYDWPPPKGC CPSKNRDTFM IQEQVALYLG ITSFKRKYPD LPRRSVDMEE RNWLQEKGLV1380
1381 SEKLCDLGIT AVWASDILDI MYADFFDKYE EYKEYIRRKH LREIEAKQKA LGLTVGAGRG1440
1441 LQARDRAMLS ATKWNAYFNK SRKDERQSCM DLQTFTINQP QPRTAPTCTR LERSTSDLIR1500
1501 AVAEEANIPA PPNLLPPRDY DEAFRNSDYA YPLTVVPDQF SMAYRQFEPA ELRAYPLDTA1560
1561 LDKPPTDLMA QLLQAKSEAV GSDEIKTSAP APKDLGQEQS AIKSVTVTAP VRRSRRSTRQ1620
1621 QTDKVRTASS SSTSSAQSVS SASSGNGSSS DTESGDESDF SSTSSCSSST GASSGAGSED1680
1681 EDGNECSSSV RLSTCGVCLR SQHRNARDMP EAFIRCYTCR KRVHPSCVDM PPRMVGRVRN1740
1741 YNWQCAGCKC CIKCRSSQRP GKMLYCEQCD RGYHIYCLGL RTVPDGRWSC ERCCFCMRCG1800
1801 ATKPEGLPQV AALSQASGGP SANGDRSKAA RNKRLKWVHE YRIDHVTKIR EHAAMFCVPC1860
1861 ARNKPAKRQS AAGAAGAAAV TPVLEATSAQ TDDSPMPSPG LTTNGGRALS PTAALSPKAA1920
1921 VPVASLPPVL EATTVTTNIA GTIGRRQAGN AVNITTMQCS SSSSSNFSGN GVTEDAANVT1980
1981 ATGTATAAAG APAATPIGIA PPPVVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
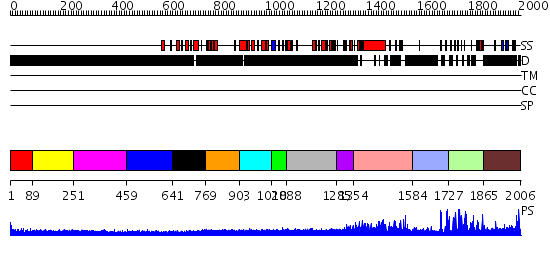
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..250] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [251..458] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [459..640] | 1.115986 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [641..768] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [769..902] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [903..1027] | 1.123987 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1028..1087] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1088..1284] | 3.109984 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1285..1353] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1354..1583] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1584..1726] | 5.045757 | Solution structure of tandem repeat of the zf-C2H2 domains of human zinc finger protein 297B | |
| 13 | View Details | [1727..1864] | 20.30103 | No description for 2ysmA was found. | |
| 14 | View Details | [1865..2006] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)