













| General Information: |
|
| Name(s) found: |
CG9485-PB /
FBpp0071526
[FlyBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1629 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glycogen biosynthetic process
[IEA]
|
| Molecular Function: |
cation binding
[IEA]
amylo-alpha-1,6-glucosidase activity [ISS] 4-alpha-glucanotransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRYQLFRWLY GLIATVDNEP LPLQIKSEEE AFGENKKKKQ LASLENAISP GKLDSVAIRT 60
61 VPSANANAMG NAGSAEIVSN QIIRRREMST MGKETESHSI PISEGQDAEH ILYRLKRGSK 120
121 LSVHPDASLL GRKIVLYTNY PAEGQKFVRT EYRVLGWQLS NGKQITSVMH PEAHVVDTDI 180
181 RSQVELNMSG TYHFYFRYLE RPDTGCSGAD GALYVQVEPT LHVGPPGAQK TIPLDSVRCQ 240
241 TVLAKLLGPL DTWEPKLRVA KEAGYNVIHF TPIQELGGSR SCYSLRDQLK VNSHFAPQKG 300
301 GKISFEDVEK VIKKCRQEWG VASICDIVLN HTANESDWLL QHPDATYSCA TCPYLRPAFL 360
361 LDATFAQCGA DIAEGSLEHV GVPAVIEQEC HLEALKYQLH TSYMSKVNIH ELYQCDVMKY 420
421 VNEFMSQVRT REPPKNVANE CRFQEIQLIQ DPQYRRLAST INFELALEIF NAFHGDCFDE 480
481 ESRFRKCAET LRRHLDALND RVRCEVQGYI NYAIDNVLAG VRYERVQGDG PRVKEISEKH 540
541 SVFMVYFTHT GTQGKSLTEI EADMYTKAGE FFMAHNGWVM GYSDPLRDFA EEQPGRANVY 600
601 LKRELISWGD SVKLRFGRRP EDSPYLWQHM TEYVQTTARI FDGVRLDNCH STPLHVAEYL 660
661 LDAARKINPE LYVVAELFTN SDYTDNVFVN RLGITSLIRE ALSAWDSHEQ GRLVYRYGGV 720
721 PVGGFQANSS RHEATSVAHA LFLDLTHDNP SPVEKRSVYD LLPSAALVSM ACCATGSNRG 780
781 YDELVPHHIH VVDEERTYQE WGKGVDSKSG IMGAKRALNL LHGQLAEEGF SQVYVDQMDP 840
841 NVVAVTRHSP ITHQSVILVA HTAFGYPSPN AGPTGIRPLR FEGVLDEIIL EASLTMQSDK 900
901 PFDRPAPFKK DPNVINGFTQ FQLNLQEHIP LAKSTVFQTQ AYSDGNNTEL NFANLRPGTV 960
961 VAIRVSMHPG PRTSFDKLQK ISAALRIGSG EEYSQLQAIV SKLDLVALSG ALFSCDDEER1020
1021 DLGKGGTAYD IPNFGKIVYC GLQGFISLLT EISPKNDLGH PLCNNLRDGN WMMDYISDRL1080
1081 TSYEDLKPLS AWFKATFEPL KNIPRYLIPC YFDAIVSGVY NVLINQVNEL MPDFIKNGHS1140
1141 FPQSLALSTL QFLSVCKSAN LPGFSPALSP PKPPKQCVTL SAGLPHFSTG YMRCWGRDTF1200
1201 IALRGSMFLT GRYNEARFII IGFGQTLRHG LIPNLLDSGS KPRFNCRDAI WWWMYCIKQY1260
1261 VEDAPKGAEI LKDKVSRIFP YDDADAHAPG AFDQLLFDVM QEALQVHFQG LQYRERNAGY1320
1321 EIDAHMVDQG FNNQIGIHPE TGFVFGGNNF NCGTWMDKMG SSQKAGNKGR PSTPRDGSAV1380
1381 ELVGLQYAVL RFMQSLAEKE VIPYTGVERK GPSGEVTKWS YKEWADRIKN NFDKYFFVSE1440
1441 SETCSVANKK LIYKDSYGAT QSWTDYQLRC NFPITLTVAP DLCNPQNAWR ALERAKKYLL1500
1501 GPLGMKTMDP EDWNYRANYD NSNDSTDCTV AHGANYHQGP EWVWPIGFYL RARLIFAKKC1560
1561 GHLDETIAET WAILRAHLRE LQTSHWRGLP ELTNDNGSYC GDSCRTQAWS VAAILEVLYD1620
1621 LHSLGADVA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
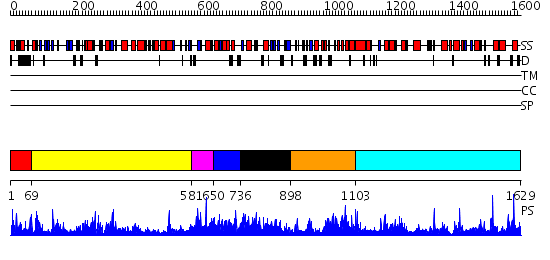
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [69..580] | 73.69897 | Neopullulanase, central domain; Neopullulanase | |
| 3 | View Details | [581..649] | 71.221849 | CRYSTAL STRUCTURE OF B. CEREUS OLIGO-1,6-GLUCOSIDASE | |
| 4 | View Details | [650..735] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [736..897] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [898..1102] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1103..1629] | 71.69897 | No description for 2jf4A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)