













| General Information: |
|
| Name(s) found: |
gi|151943580
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 960 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDLETVAKF LAESVIASTA KTSERNLRQL ETQDGFGLTL LHVIASTNLP LSTRLAGALF 60
61 FKNFIKRKWV DENGNHLLPA NNVELIKKEI VPLMISLPNN LQVQIGEAIS SIADSDFPDR 120
121 WPTLLSDLAS RLSNDDMVTN KGVLTVAHSI FKRWRPLFRS DELFLEIKLV LDVFTAPFLN 180
181 LLKTVDEQIT ANENNKASLN ILFDVLLVLI KLYYDFNCQD IPEFFEDNIQ VGMGIFHKYL 240
241 SYSNPLLEDP DETEHASVLI KVKSSIQELV QLYTTRYEDV FGPMINEFIQ ITWNLLTSIS 300
301 NQPKYDILVS KSLSFLTAVT RIPKYFEIFN NESAMNNITE QIILPNVTLR EEDVELFEDD 360
361 PIEYIRRDLE GSDTDTRRRA CTDFLKELKE KNEVLVTNIF LAHMKGFVDQ YMSDPSKNWK 420
421 FKDLYIYLFT ALAINGNITN AGVSSTNNLL NVVDFFTKEI APDLTSNNIP HIILRVDAIK 480
481 YIYTFRNQLT KAQLIELMPI LATFLQTDEY VVYTYAAITI EKILTIRESN TSPAFIFHKE 540
541 DISNSTEILL KNLIALILKH GSSPEKLAEN EFLMRSIFRV LQTSEDSIQP LFPQLLAQFI 600
601 EIVTIMAKNP SNPRFTHYTF ESIGAILNYT QRQNLPLLVD SMMPTFLTVF SEDIQEFIPY 660
661 VFQIIAFVVE QSATIPESIK PLAQPLLAPN VWELKGNIPA VTRLLKSFIK TDSSIFPDLV 720
721 PVLGIFQRLI ASKAYEVHGF DLLEHIMLLI DMNRLRPYIK QIAVLLLQRL QNSKTERYVK 780
781 KLTVFFGLIS NKLGSDFLIH FIDEVQDGLF QQIWGNFIIT TLPTIGNLLD RKIALIGVLN 840
841 MVINGQFFQS KYPTLISSTM NSIIETASSQ SIANLKNDYV DLDNLEEIST FGSHFSKLVS 900
901 ISEKPFDPLP EIDVNNGVRL YVAEALNKYN AISGNTFLNT ILPQLTQENQ VKLNQLLVGN 960
961 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
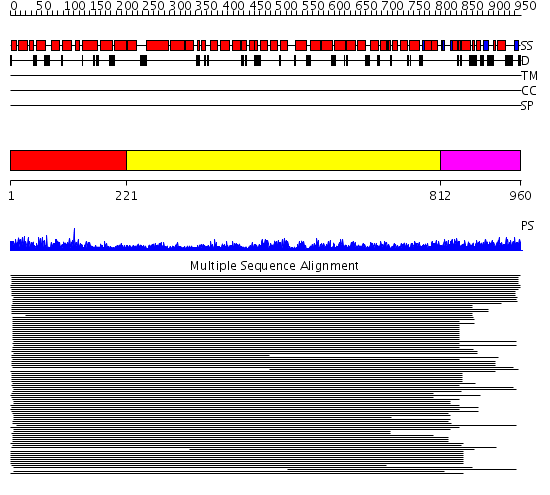
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..220] | 6.154993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [221..811] | 18.235994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [812..960] | 1.094981 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)