













| General Information: |
|
| Name(s) found: |
GBB1_RAT
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Rattus norvegicus |
| Length: | 340 amino acids |
Gene Ontology: |
|
| Cellular Component: |
heterotrimeric G-protein complex
[UNKNOWN][IMP][TAS]
plasma membrane [UNKNOWN] photoreceptor outer segment [UNKNOWN][IEA] |
| Biological Process: |
cell proliferation
[UNKNOWN][IEA]
sensory perception of taste [UNKNOWN][IEA] G-protein coupled receptor protein signaling pathway [UNKNOWN][IMP][TAS] signal transduction [IEA] activation of phospholipase C activity by G-protein coupled receptor protein signaling pathway coupled to IP3 second messenger [UNKNOWN][IEA] elevation of cytosolic calcium ion concentration [IMP] hormone-mediated signaling pathway [UNKNOWN] |
| Molecular Function: |
G-protein-coupled receptor binding
[IMP]
type 1 angiotensin receptor binding [IMP] GTPase activity [UNKNOWN][IEA] signal transducer activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA 60
61 MHWGTDSRLL VSASQDGKLI IWDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI 120
121 CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF 180
181 TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TFTGHESDIN AICFFPNGNA 240
241 FATGSDDATC RLFDLRADQE LMTYSHDNII CGITSVSFSK SGRLLLAGYD DFNCNVWDAL 300
301 KADRAGVLAG HDNRVSCLGV TDDGMAVATG SWDSFLKIWN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
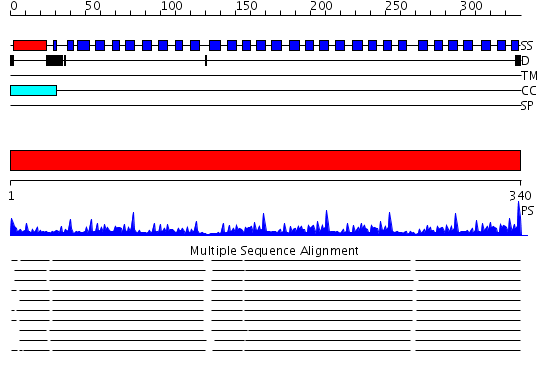
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..340] | 76.09691 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)