













| General Information: |
|
| Name(s) found: |
mtm-6 /
CE37116
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
phospholipid dephosphorylation
[IEA protein amino acid dephosphorylation [IEA endocytosis [IMP dephosphorylation [IEA positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
zinc ion binding
[IEA inositol or phosphatidylinositol phosphatase activity [IEA protein tyrosine phosphatase activity [IEA phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFEDIGISK VDKVCLVDRL GCQENLVGTV HVTTTHIIFR AENGSKELWL ATGLISSVEK 60
61 GTLTAAGCML VIRCKHFQVI TLLISRDKSC QDLYETLQRA AKPVNVTELL AFENREPVED 120
121 VRGWRRLDWN SEMTRQGITK SQWTESNINE GYTICDTYPN KLWFPTAAST SVLLGSCKFR 180
181 SRGRLPVLTY FHQQTEAALC RCAQPLTGFS ARCVEDEKLM ELVGKANTNS DNLFLVDTRP 240
241 RVNAMVNKVQ GKGFEDERNY SNMRFHFFDI ENIHVMRASQ ARLLDAVTKC RDVTEYWKTL 300
301 EASGWLKHVR SVVECSLFLA ESISRGTSCV VHCSDGWDRT SQVVALCQLL LDPYYRTIHG 360
361 FQVLIEKDWL GFGHKFDDRC GHVGALNDEA GKEVSPIFTQ WLDCIWQIMQ QKPRAFQFNE 420
421 RYLIEMHEHV YSCQFGTFIG NCDKDRRDLN LSKRTKSLWT WMDARHDDYM NPFYSPTAHV 480
481 ALLDLDTRAA RFTVWTAMYN RFDNGLQPRE RLEDLTMAAM EHVGVLESHV AQLRTRLAEL 540
541 KTQQNQQITS TNTPTNMVDS GMSSATDDLK NLSLSHPLDP LSSTLPILER ATSQESGVMD 600
601 SSLYYPDEAL TKYSLKWQPL RGADRCSNPA CRGEFSSTIE RRIHCHLCGM IFCRRCLKVS 660
661 ADERERVCDK CKTD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
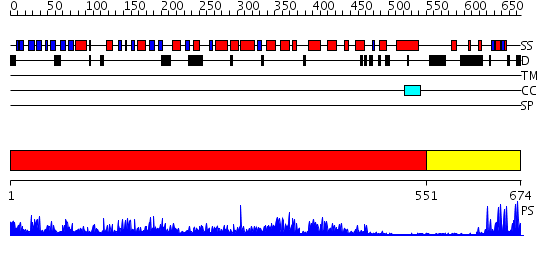
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..550] | 177.0 | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | |
| 2 | View Details | [551..674] | 15.0 | Hrs |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)