













| General Information: |
|
| Name(s) found: |
2AAB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to cadmium ion
[IEP]
regulation of phosphorylation [ISS] |
| Molecular Function: |
protein phosphatase type 2A regulator activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMIDEPLYP IAVLIDELKN DDIQLRLNSI RRLSTIARAL GEERTRKELI PFLSENNDDD 60
61 DEVLLAMAEE LGVFIPYVGG VEYAHVLLPP LETLSTVEET CVREKAVESL CRVGSQMRES 120
121 DLVDHFISLV KRLAAGEWFT ARVSACGVFH IAYPSAPDML KTELRSLYTQ LCQDDMPMVR 180
181 RAAATNLGKF AATVESAHLK TDVMSMFEDL TQDDQDSVRL LAVEGCAALG KLLEPQDCVQ 240
241 HILPVIVNFS QDKSWRVRYM VANQLYELCE AVGPEPTRTE LVPAYVRLLR DNEAEVRIAA 300
301 AGKVTKFCRI LNPEIAIQHI LPCVKELSSD SSQHVRSALA SVIMGMAPVL GKDATIEHLL 360
361 PIFLSLLKDE FPDVRLNIIS KLDQVNQVIG IDLLSQSLLP AIVELAEDRH WRVRLAIIEY 420
421 IPLLASQLGV GFFDDKLGAL CMQWLQDKVH SIRDAAANNL KRLAEEFGPE WAMQHIVPQV 480
481 LEMVNNPHYL YRMTILRAVS LLAPVMGSEI TCSKLLPVVM TASKDRVPNI KFNVAKVLQS 540
541 LIPIVDQSVV EKTIRPGLVE LSEDPDVDVR FFANQALQSI DNVMMSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
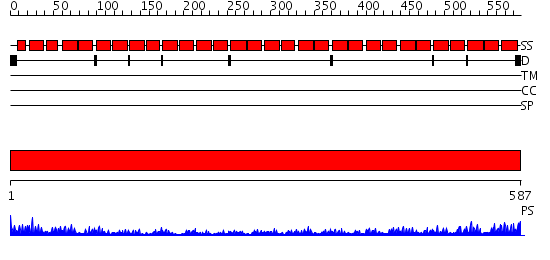
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..587] | 102.0 | Constant regulatory domain of protein phosphatase 2a, pr65alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)