













| General Information: |
|
| Name(s) found: |
PNP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 991 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
RNA processing
[IEA]
mRNA catabolic process [IEA] |
| Molecular Function: |
nucleic acid binding
[ISS]
3'-5'-exoribonuclease activity [IEA][ISS] RNA binding [IEA][ISS] polyribonucleotide nucleotidyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSIVNRASS ASLPNFLAWR ALGFRTICSG RLGFAPSVPD SPVSAGTKIL ESFKEEFEVG 60
61 SRVVSFETGK IARFANGSVV LGMDETKVLS TVTCAKTDSP RDFLPLTVDY QEKQYAQGLI 120
121 PNTYMRREGA PKERELLCGR LIDRPIRPLF PTGFYHEVQI MASVLSSDGK QDPDILAANA 180
181 SSAALMLSDV PWGGPIGVIR IGRICGQFVV NPTMDELSSS DLNLIYACTR DKTMMIDVQS 240
241 REISEKDLAA ALRLAHPEAV KYLDPQIRLA EKAGKQKKEY KLSMLSDKTL EKVADLAATR 300
301 IESVFTDPSY GKFERGEALD NIGKDVRKVF EEEGDQESLS ILPKAVDTVR KKVVRSRMIS 360
361 DGFRVDGRHV DEVRPIYCES HYLPALHGSA LFSRGDTQVL CTVTLGAPAE AQSLDSLVGP 420
421 PKKRFMLHYS FPPYCTNEVG KRGGLNRREV GHGTLAEKAL LAVLPPEEAF PYTIRINSEV 480
481 MSSDGSTSMA SVCGGSMALM DAGIPLRAHV AGVSVGLITD VDPSSGEIKD YRIVTDILGL 540
541 EDHLGDMDFK IAGTRDGVTA IQLDIKPAGI PLDIVCESLE NAREARLQIL DHMERNINSP 600
601 RGQDGAYSPR LATLKYSNDS LRTLIGPMGV LKRKIEVETG ARLSIDNGTL TIVAKNQDVM 660
661 EKAQEQVDFI IGRELVVGGV YKGTVSSIKE YGAFVEFPGG QQGLLHMSEL SHEPVSKVSD 720
721 VLDIGQCITT MCIETDVRGN IKLSRKALLP KPKRKPASDA GKDPVMKESS TVYIENSSVG 780
781 EIVASMPSIV TPLQKSRLSV PAVVIRTAVE CNEAEKSSPV NDNDKPRRAA TSKPDRKPKS 840
841 TASKLIATQK EEEALESIAP EETSAECGEI LKQDGKLKSV SPKNNSTASN LVSFSKAKKS 900
901 TMKENLSENK AEESASVSTR KLKIGTEMTA TVDHVRALGL VLDLGGEIRG MYIFQGDKDK 960
961 FKKGDTLRVK CTSFNTKGVP VMALVDEEGE E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
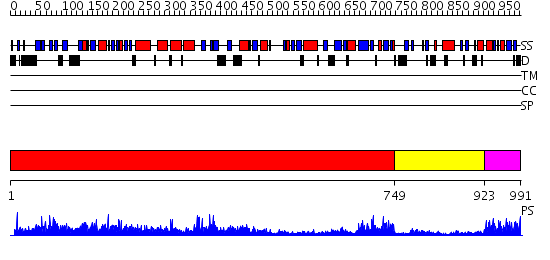
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..748] | 154.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [749..922] | 16.0 | Catalytic domain of E. coli RNase E | |
| 3 | View Details | [923..991] | 3.69897 | Solution structure of the S1 RNA binding domain from human hypothetical protein BAA11502 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|