













| General Information: |
|
| Name(s) found: |
CUL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 742 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast envelope
[IDA]
SCF ubiquitin ligase complex [ISS] |
| Biological Process: |
cell cycle
[ISS]
ubiquitin-dependent protein catabolic process [IEA] |
| Molecular Function: |
ubiquitin protein ligase binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKDSVLEA GWSVMEAGVA KLQKILEEVP DEPPFDPVQR MQLYTTVHNL CTQKPPNDYS 60
61 QQIYDRYGGV YVDYNKQTVL PAIREKHGEY MLRELVKRWA NQKILVRWLS HFFEYLDRFY 120
121 TRRGSHPTLS AVGFISFRDL VYQELQSKAK DAVLALIHKE REGEQIDRAL LKNVIDVYCG 180
181 NGMGELVKYE EDFESFLLED SASYYSRNAS RWNQENSCPD YMIKAEESLR LEKERVTNYL 240
241 HSTTEPKLVA KVQNELLVVV AKQLIENEHS GCRALLRDDK MDDLARMYRL YHPIPQGLDP 300
301 VADLFKQHIT VEGSALIKQA TEAATDKAAS TSGLKVQDQV LIRQLIDLHD KFMVYVDECF 360
361 QKHSLFHKAL KEAFEVFCNK TVAGVSSAEI LATYCDNILK TGGGIEKLEN EDLELTLEKV 420
421 VKLLVYISDK DLFAEFFRKK QARRLLFDRN GNDYHERSLL TKFKELLGAQ FTSKMEGMLT 480
481 DMTLAKEHQT NFVEFLSVNK TKKLGMDFTV TVLTTGFWPS YKTTDLNLPI EMVNCVEAFK 540
541 AYYGTKTNSR RLSWIYSLGT CQLAGKFDKK TIEIVVTTYQ AAVLLLFNNT ERLSYTEILE 600
601 QLNLGHEDLA RLLHSLSCLK YKILIKEPMS RNISNTDTFE FNSKFTDKMR RIRVPLPPMD 660
661 ERKKIVEDVD KDRRYAIDAA LVRIMKSRKV LGHQQLVSEC VEHLSKMFKP DIKMIKKRIE 720
721 DLISRDYLER DTDNPNTFKY LA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
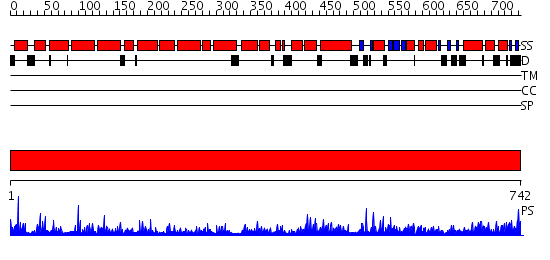
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..742] | 1000.0 | No description for 2hyeC was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)