













| General Information: |
|
| Name(s) found: |
SMC4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1241 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
chromosome [IEA] |
| Biological Process: |
chromosome organization
[IEA]
chromosome segregation [ISS] |
| Molecular Function: |
ATP binding
[ISS]
transporter activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEDEPMGGG ESEPEQRKSG TPRLYIKELV MRNFKSYAGE QRVGPFHKSF SAVVGPNGSG 60
61 KSNVIDAMLF VFGKRAKQMR LNKVSELIHN STNHQNLDSA GVSVQFEEII DLENGLYETV 120
121 PGSDFMITRV AFRDNSSKYY INERSSNFTE VTKKLKGKGV DLDNNRFLIL QGEVEQISLM 180
181 KPKAQGPHDE GFLEYLEDII GTNKYVEKID ELNKQLETLN ESRSGVVQMV KLAEKERDNL 240
241 EGLKDEAETY MLKELSHLKW QEKATKMAYE DTVAKITEQR DSLQNLENSL KDERVKMDES 300
301 NEELKKFESV HEKHKKRQEV LDNELRACKE KFKEFERQDV KHREDLKHVK QKIKKLEDKL 360
361 EKDSSKIGDM TKESEDSSNL IPKLQENIPK LQKVLLDEEK KLEEIKAIAK VETEGYRSEL 420
421 TKIRAELEPW EKDLIVHRGK LDVASSESEL LSKKHEAALK AFTDAQKQLS DISTRKKEKA 480
481 AATTSWKADI KKKKQEAIEA RKVEEESLKE QETLVPQEQA AREKVAELKS AMNSEKSQNE 540
541 VLKAVLRAKE NNQIEGIYGR MGDLGAIDAK YDVAISTACA GLDYIVVETT SSAQACVELL 600
601 RKGNLGFATF MILEKQTDHI HKLKEKVKTP EDVPRLFDLV RVKDERMKLA FYAALGNTVV 660
661 AKDLDQATRI AYGGNREFRR VVALDGALFE KSGTMSGGGG KARGGRMGTS IRATGVSGEA 720
721 VANAENELSK IVDMLNNIRE KVGNAVRQYR AAENEVSGLE MELAKSQREI ESLNSEHNYL 780
781 EKQLASLEAA SQPKTDEIDR LKELKKIISK EEKEIENLEK GSKQLKDKLQ TNIENAGGEK 840
841 LKGQKAKVEK IQTDIDKNNT EINRCNVQIE TNQKLIKKLT KGIEEATREK ERLEGEKENL 900
901 HVTFKDITQK AFEIQETYKK TQQLIDEHKD VLTGAKSDYE NLKKSVDELK ASRVDAEFKV 960
961 QDMKKKYNEL EMREKGYKKK LNDLQIAFTK HMEQIQKDLV DPDKLQATLM DNNLNEACDL1020
1021 KRALEMVALL EAQLKELNPN LDSIAEYRSK VELYNGRVDE LNSVTQERDD TRKQYDELRK1080
1081 RRLDEFMAGF NTISLKLKEM YQMITLGGDA ELELVDSLDP FSEGVVFSVR PPKKSWKNIA1140
1141 NLSGGEKTLS SLALVFALHH YKPTPLYVMD EIDAALDFKN VSIVGHYVKD RTKDAQFIII1200
1201 SLRNNMFELA DRLVGIYKTD NCTKSITINP GSFAVCQKTP A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
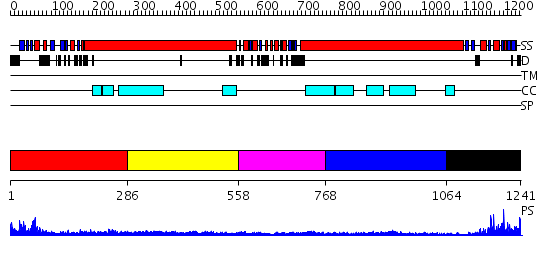
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..285] | 23.30103 | Sc Smc1hd:Scc1-C complex, ATPgS | |
| 2 | View Details | [286..557] | 2.85 | Tropomyosin | |
| 3 | View Details | [558..767] | 4.46 | Smc hinge domain | |
| 4 | View Details | [768..1063] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1064..1241] | 17.0 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)