













| General Information: |
|
| Name(s) found: |
PEP_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 495 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
gynoecium development
[IMP]
shoot development [IMP] |
| Molecular Function: |
nucleic acid binding
[ISS]
RNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVADSVEN NGSINLPENE NLIPAGFSAA ALLDENSGAF PELNQPDSLA AAETTFPDTN 60
61 DSAEERWPGW PGDCVFRMIV PVTKVGAIIG RKGDFIKKMC EETRARIKVL DGPVNTPDRI 120
121 VLISGKEEPE AYMSPAMDAV LRVFRRVSGL PDNDDDDVQN AGSVFSSVRL LVASTQAINL 180
181 IGKQGSLIKS IVENSGASVR ILSEEETPFY AAQDERIVDL QGEALKILKA LEAIVGHLRR 240
241 FLVDHTVVPL FEKQYLARVS QTRQEEPLAE SKSSLHTISS NLMEPDFSLL ARREPLFLER 300
301 DSRVDSRVQP SGVSIYSQDP VLSARHSPGL ARVSSAFVTQ VSQTMQIPFS YAEDIIGVEG 360
361 ANIAYIRRRS GATITIKESP HPDQITVEIK GTTSQVQTAE QLIQEFIINH KEPVSVSGGY 420
421 ARIDSGYVPA YPPQLSNRQE PLPSTYMGTE PVQYRPTAYS QLGGPSTYTP TLTGQTYGSE 480
481 YRPASDVGGY SSYNL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
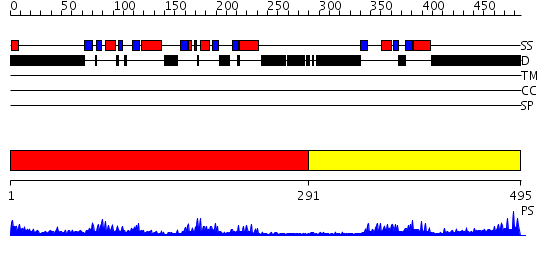
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..290] | 33.39794 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [291..495] | 12.09691 | No description for 2hh2A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)