













| General Information: |
|
| Name(s) found: |
gi|15230754
[NCBI NR]
gi|12322678 [NCBI NR] gi|12321919 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[IEA][ISS]
two-component signal transduction system (phosphorelay) [IEA] regulation of transcription, DNA-dependent [IEA] protein amino acid phosphorylation [IEA] |
| Molecular Function: |
two-component sensor activity
[IEA]
ATP binding [IEA] protein kinase activity [IEA][ISS] protein serine/threonine kinase activity [IEA] signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEELLKKML ELEQSQELLK QEMSRLKLST ELRQPSHPVL PRRPLRRIQG SMNSNPSPGK 60
61 FTDKQYLNIL QSLAQSVHVL DLNTRIIFWN AMSEKLYGYS AAEVVGRNPV HVIVDDQNAA 120
121 FALNVARRCA NGESWTGEFP VKTKSGKIFS AVTTCSPFYD DNGTVVGIIS ITSDIAPYLN 180
181 PRLSLPRLKP QEPERKLGLD SKGAVISKPG LDSDQPIQVS IASKISSLAS KLSNKVRSKM 240
241 RAGDNSACGD SHHSDHDVFG DTLSDHRDDA ASSGASTPRG DFIQSPFGVF TCYDDKFPSK 300
301 PSKDSSDRKP AIHKVPTSKA EEWMVKKGLS RPWKGNEQEG SRVRPTHSVW SWVENEQEKD 360
361 KYHQIYPSAG VKSESHGSES NKPTDDEASN MWSSSINANS TNSASSCGST SRSVMDKVDI 420
421 DSDPLEHEIL WDDLTIGEQI GRGSCGTVYH GIWFGSDVAV KVFSKQEYSE SVIKSFEKEV 480
481 SLMKRLRHPN VLLFMGAVTS PQRLCIVSEF VPRGSLFRLL QRSMSKLDWR RRINMALDIA 540
541 RGMNYLHCCS PPIIHRDLKS SNLLVDRNWT VKVADFGLSR IKHQTYLTSK SGKGTPQWMA 600
601 PEVLRNESAD EKSDIYSFGV VLWELATEKI PWENLNSMQV IGAVGFMNQR LEIPKDTDPD 660
661 WISLIESCWH R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
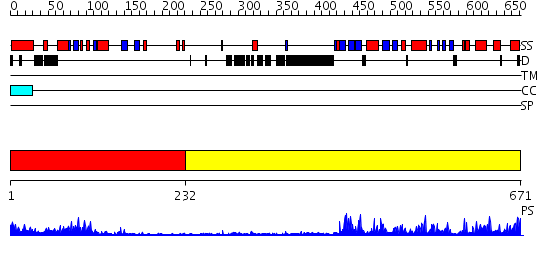
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..231] | 14.39794 | No description for 2gj3A was found. | |
| 2 | View Details | [232..671] | 89.39794 | No description for 2h8hA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)