













| General Information: |
|
| Name(s) found: |
PUB53_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 819 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
protein ubiquitination [IEA] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA]
ATP binding [IEA] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSFFKNSYL DPKAVRAQLP KRADLAAPSE PMTVALAISG SIKSKNVIKW ALNKFGSDKN 60
61 VTFKLIHIHP KITTLPTASG NIVSISEELE EVAAAYRQKV MQETKETLLK PFKKMCERKK 120
121 LKIDETRFES SLTKVAVELQ VLESNSVAVA ITKEVNQHLI SNLIIGRSSQ AASSRNYDIT 180
181 ASISASVSNL CTVYVVSNGG VHILAKDTSD TERNDTSIES GFERTSSSCS SGSGANSDVM 240
241 SNALKSNPHT LSNKRMQNLP TIVRGVSVPM ETSSTESDET KKRSSDAAEE ASKRSSPETS 300
301 RSVSWNPQFR DFDERKDAMS SMSSNFEYGN VVTPLGHYFT DNQDTLNEIS KLRAELRHAH 360
361 EMYAVAQVET LDASRKLNEL KFEELTLLEH ETKGIAKKET EKFEQKRREE REAAQRREAE 420
421 MKATHEAKEK EKLEESSLVA PKLQYQEFTW EEIINATSSF SEDLKIGMGA YGDVYKCNLH 480
481 HTIAAVKVLH SAESSLSKQF DQELEILSKI RHPHLVLLLG ACPDHGALVY EYMENGSLED 540
541 RLFQVNDSQP IPWFVRLRIA WEVASALVFL HKSKPTPIIH RDLKPANILL NHNFVSKVGD 600
601 VGLSTMIQAA DPLSTKFTMY KQTSPVGTLC YIDPEYQRTG RISPKSDVYA FGMIILQLLT 660
661 GQQAMALTYT VETAMENNND DELIQILDEK AGNWPIEETR QLAALALQCT ELRSKDRPDL 720
721 EDQILPVLES LKKVADKARN SLSAAPSQPP SHFFCPLLKD VMKEPCIAAD GYTYDRRAIE 780
781 EWMENHRTSP VTNSPLQNVN LLPNHTLYAA IVEWRNRNQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
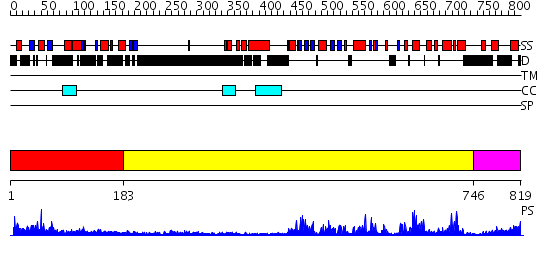
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 4.18 | "Hypothetical" protein MJ0577 | |
| 2 | View Details | [183..745] | 90.69897 | No description for 2h8hA was found. | |
| 3 | View Details | [746..819] | 58.0 | No description for 2qoqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)