













| General Information: |
|
| Name(s) found: |
FAS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 487 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin assembly complex
[IPI]
|
| Biological Process: |
cell proliferation
[IMP]
heterochromatin formation [IMP] leaf development [IMP] double-strand break repair via homologous recombination [IMP] DNA recombination [IMP] pollen development [IGI] meristem structural organization [IMP] nucleosome assembly [IDA] trichome differentiation [IMP] |
| Molecular Function: |
nucleotide binding
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGGTIQISW HDGKPVLTVD FHPISGLLAT AGADYDIKLW LINSGQAEKK VPSVSYQSSL 60
61 TYHGCAVNTI RFSPSGELLA SGADGGELFI WKLHPSETNQ SWKVHKSLSF HRKDVLDLQW 120
121 SPDDAYLISG SVDNSCIIWD VNKGSVHQIL DAHCHYVQGV AWDPLAKYVA SLSSDRTCRI 180
181 YANKPQTKSK GVEKMNYVCQ HVIMKADQQR GDETKTIKTH LFHDETLPSF FRRLSWSPDG 240
241 SFLLIPAGSF KVSPTSEAVN ATYVFSRKDL SRPALQLPGA SKPVVVVRFC PVAFKLRGSS 300
301 SEEGFFKLPY RLVFAIATLN SVYIYDTECV APIAVLAGLH YAAITDITWS PNASYLALSS 360
361 QDGYCTLVEF EDKELGEAVS ISVGKKPVDG EEKKHDLEKG DELMTETTPD ESKKQAELEQ 420
421 NEESKQPLPS KITTDGKEKE HIMQKTDDEV MTETRHEEEN QPLQSKVNTP VSNKPARKRI 480
481 TPMAIDP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
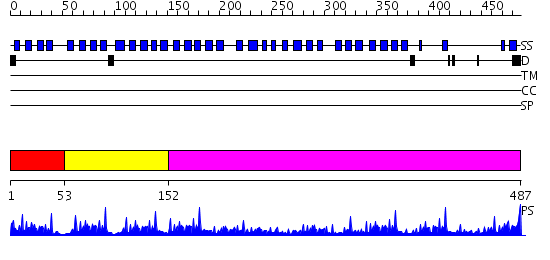
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..52] | 70.69897 | No description for 2gnqA was found. | |
| 2 | View Details | [53..151] | 4.39794 | Crystal Structure of human dipeptidyl peptidase IV in complex with a decapeptide (tNPY) at 2.3 Ang. Resolution | |
| 3 | View Details | [152..487] | 10.0 | No description for 2i3sA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)