













| General Information: |
|
| Name(s) found: |
CUL3A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 732 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[TAS]
[NOT]SCF ubiquitin ligase complex [IPI] |
| Biological Process: |
response to red or far red light
[IMP]
embryonic development ending in seed dormancy [IGI] endosperm development [IGI] positive regulation of flower development [IMP] cell cycle [ISS] ubiquitin-dependent protein catabolic process [TAS] |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNQKKRNFQ IEAFKHRVVV DPKYADKTWQ ILERAIHQIY NQDASGLSFE ELYRNAYNMV 60
61 LHKFGEKLYT GFIATMTSHL KEKSKLIEAA QGGSFLEELN KKWNEHNKAL EMIRDILMYM 120
121 DRTYIESTKK THVHPMGLNL WRDNVVHFTK IHTRLLNTLL DLVQKERIGE VIDRGLMRNV 180
181 IKMFMDLGES VYQEDFEKPF LDASSEFYKV ESQEFIESCD CGDYLKKSEK RLTEEIERVA 240
241 HYLDAKSEEK ITSVVEKEMI ANHMQRLVHM ENSGLVNMLL NDKYEDLGRM YNLFRRVTNG 300
301 LVTVRDVMTS HLREMGKQLV TDPEKSKDPV EFVQRLLDER DKYDKIINTA FGNDKTFQNA 360
361 LNSSFEYFIN LNARSPEFIS LFVDDKLRKG LKGITDVDVE VILDKVMMLF RYLQEKDVFE 420
421 KYYKQHLAKR LLSGKTVSDD AERSLIVKLK TECGYQFTSK LEGMFTDMKT SEDTMRGFYG 480
481 SHPELSEGPT LIVQVLTTGS WPTQPAVPCN LPAEVSVLCE KFRSYYLGTH TGRRLSWQTN 540
541 MGTADIKAIF GKGQKHELNV STFQMCVLML FNNSDRLSYK EIEQATEIPA ADLKRCLQSL 600
601 ACVKGKNVIK KEPMSKDIGE EDLFVVNDKF TSKFYKVKIG TVVAQKETEP EKQETRQRVE 660
661 EDRKPQIEAA IVRIMKSRKI LDHNNIIAEV TKQLQPRFLA NPTEIKKRIE SLIERDFLER 720
721 DSTDRKLYRY LA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
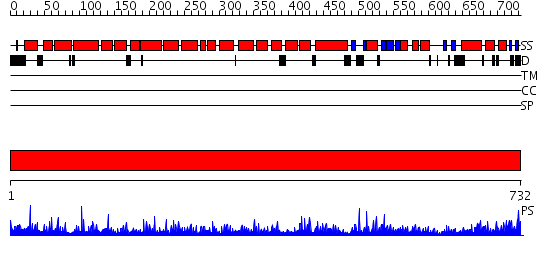
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..732] | 1000.0 | No description for 2hyeC was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)