













| General Information: |
|
| Name(s) found: |
CDPKX_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 521 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA][ISS]
|
| Molecular Function: |
ATP binding
[IEA]
calcium ion binding [IEA] protein kinase activity [IEA] calmodulin-dependent protein kinase activity [ISS] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNCLAKKYG LVMKPQQNGE RSVEIENRRR STHQDPSKIS TGTNQPPPWR NPAKHSGAAA 60
61 ILEKPYEDVK LFYTLSKELG RGQFGVTYLC TEKSTGKRFA CKSISKKKLV TKGDKEDMRR 120
121 EIQIMQHLSG QPNIVEFKGA YEDEKAVNLV MELCAGGELF DRILAKGHYS ERAAASVCRQ 180
181 IVNVVNICHF MGVMHRDLKP ENFLLSSKDE KALIKATDFG LSVFIEEGRV YKDIVGSAYY 240
241 VAPEVLKRRY GKEIDIWSAG IILYILLSGV PPFWAETEKG IFDAILEGEI DFESQPWPSI 300
301 SNSAKDLVRR MLTQDPKRRI SAAEVLKHPW LREGGEASDK PIDSAVLSRM KQFRAMNKLK 360
361 KLALKVIAEN IDTEEIQGLK AMFANIDTDN SGTITYEELK EGLAKLGSRL TEAEVKQLMD 420
421 AADVDGNGSI DYIEFITATM HRHRLESNEN VYKAFQHFDK DGSGYITTDE LEAALKEYGM 480
481 GDDATIKEIL SDVDADNDGR INYDEFCAMM RSGNPQQPRL F |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
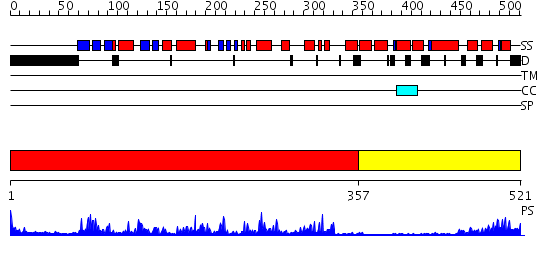
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..356] | 72.0 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 2 | View Details | [357..521] | 25.0 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)