













| General Information: |
|
| Name(s) found: |
CDPKO_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA][ISS]
|
| Molecular Function: |
ATP binding
[IEA]
calcium ion binding [IEA] calmodulin-dependent protein kinase activity [ISS] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSCVSSPLK GSPFGKRPVR RRHSSNSRTS SVPRFDSSTN LSRRLIFQPP SRVLPEPIGD 60
61 GIHLKYDLGK ELGRGEFGVT HECIEISTRE RFACKRISKE KLRTEIDVED VRREVEIMRC 120
121 LPKHPNIVSF KEAFEDKDAV YLVMEICEGG ELFDRIVSRG HYTERAAASV AKTILEVVKV 180
181 CHEHGVIHRD LKPENFLFSN GTETAQLKAI DFGLSIFFKP AQRFNEIVGS PYYMAPEVLR 240
241 RNYGPEIDVW SAGVILYILL CGVPPFWAET EEGIAHAIVR GNIDFERDPW PKVSHEAKEL 300
301 VKNMLDANPY SRLTVQEVLE HPWIRNAERA PNVNLGDNVR TKIQQFLLMN RFKKKVLRIV 360
361 ADNLPNEEIA AIVQMFQTMD TDKNGHLTFE ELRDGLKKIG QVVPDGDVKM LMDAADTDGN 420
421 GMLSCDEFVT LSIHLKRMGC DEHLQEAFKY FDKNGNGFIE LDELKVALCD DKLGHANGND 480
481 QWIKDIFFDV DLNKDGRISF DEFKAMMKSG TDWKMASRQY SRALLNALSI KMFKEDFGDN 540
541 GPKSHSMEFP IARKRAKLLD APKNKSMELQ ISKTYKPSGL RN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
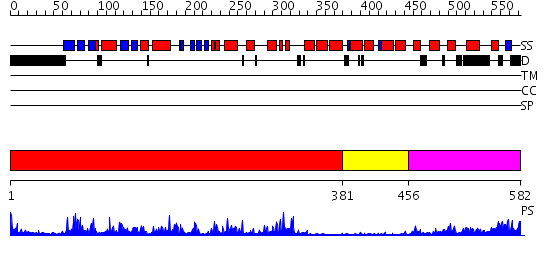
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..380] | 73.09691 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 2 | View Details | [381..455] | 3.22 | Calcium vector protein | |
| 3 | View Details | [456..582] | 24.0 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)