













| General Information: |
|
| Name(s) found: |
CPNB1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 600 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA][NAS] cytosolic ribosome [IDA] stromule [IDA] membrane [IDA] apoplast [IDA] chloroplast envelope [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IEP]
cell death [IMP] systemic acquired resistance [IMP] chaperone mediated protein folding requiring cofactor [IDA] protein folding [TAS] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTFTATSS IGSMVAPNGH KSDKKLISKL SSSSFGRRQS VCPRPRRSSS AIVCAAKELH 60
61 FNKDGTTIRR LQAGVNKLAD LVGVTLGPKG RNVVLESKYG SPRIVNDGVT VAREVELEDP 120
121 VENIGAKLVR QAAAKTNDLA GDGTTTSVVL AQGFIAEGVK VVAAGANPVL ITRGIEKTAK 180
181 ALVTELKKMS KEVEDSELAD VAAVSAGNND EIGNMIAEAM SKVGRKGVVT LEEGKSAENN 240
241 LYVVEGMQFD RGYISPYFVT DSEKMSVEFD NCKLLLVDKK ITNARDLVGV LEDAIRGGYP 300
301 ILIIAEDIEQ EALATLVVNK LRGTLKIAAL RAPGFGERKS QYLDDIAILT GATVIREEVG 360
361 LSLDKAGKEV LGNASKVVLT KETSTIVGDG STQDAVKKRV TQIKNLIEQA EQDYEKEKLN 420
421 ERIAKLSGGV AVIQVGAQTE TELKEKKLRV EDALNATKAA VEEGIVVGGG CTLLRLASKV 480
481 DAIKATLDND EEKVGADIVK RALSYPLKLI AKNAGVNGSV VSEKVLSNDN VKFGYNAATG 540
541 KYEDLMAAGI IDPTKVVRCC LEHAASVAKT FLMSDCVVVE IKEPEPVPVG NPMDNSGYGY 600
601 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
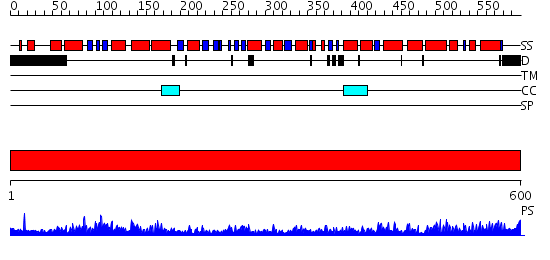
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..600] | 167.0 | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)