













| General Information: |
|
| Name(s) found: |
HDA19_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 501 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
|
| Biological Process: |
DNA mediated transformation
[IMP]
negative regulation of transcription [IDA] histone acetylation [IDA] jasmonic acid and ethylene-dependent systemic resistance [IMP] histone deacetylation [ISS] |
| Molecular Function: |
protein binding
[IPI]
histone deacetylase activity [IDA][IMP][ISS] basal transcription repressor activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTGGNSLAS GPDGVKRKVC YFYDPEVGNY YYGQGHPMKP HRIRMTHALL AHYGLLQHMQ 60
61 VLKPFPARDR DLCRFHADDY VSFLRSITPE TQQDQIRQLK RFNVGEDCPV FDGLYSFCQT 120
121 YAGGSVGGSV KLNHGLCDIA INWAGGLHHA KKCEASGFCY VNDIVLAILE LLKQHERVLY 180
181 VDIDIHHGDG VEEAFYATDR VMTVSFHKFG DYFPGTGHIQ DIGYGSGKYY SLNVPLDDGI 240
241 DDESYHLLFK PIMGKVMEIF RPGAVVLQCG ADSLSGDRLG CFNLSIKGHA ECVKFMRSFN 300
301 VPLLLLGGGG YTIRNVARCW CYETGVALGV EVEDKMPEHE YYEYFGPDYT LHVAPSNMEN 360
361 KNSRQMLEEI RNDLLHNLSK LQHAPSVPFQ ERPPDTETPE VDEDQEDGDK RWDPDSDMDV 420
421 DDDRKPIPSR VKREAVEPDT KDKDGLKGIM ERGKGCEVEV DESGSTKVTG VNPVGVEEAS 480
481 VKMEEEGTNK GGAEQAFPPK T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
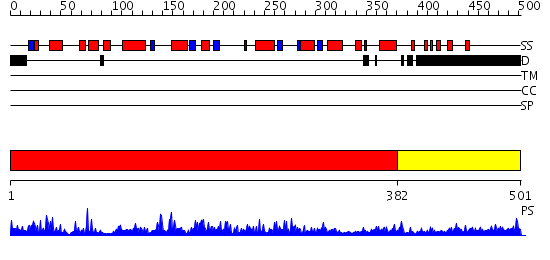
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..381] | 97.69897 | Crystal Structure of human HDAC8 complexed with Trichostatin A | |
| 2 | View Details | [382..501] | 4.122996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)