













| General Information: |
|
| Name(s) found: |
GLR28_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 947 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
endomembrane system [IEA] |
| Biological Process: |
response to light stimulus
[NAS]
cellular calcium ion homeostasis [NAS] |
| Molecular Function: |
intracellular ligand-gated ion channel activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPKKNNNTF LSYFVCLFLL LEVGLGQNQI SEIKVGVVLD LNTTFSKICL TSINLALSDF 60
61 YKDHPNYRTR LALHVRDSMK DTVQASAAAL DLIQNEQVSA IIGPIDSMQA KFMIKLANKT 120
121 QVPTISFSAT SPLLTSIKSD YFVRGTIDDS YQVKAIAAIF ESFGWRSVVA IYVDNELGEG 180
181 IMPYLFDALQ DVQVDRSVIP SEANDDQILK ELYKLMTRQT RVFVVHMASR LASRIFEKAT 240
241 EIGMMEEGYV WLMTNGMTHM MRHIHHGRSL NTIDGVLGVR SHVPKSKGLE DFRLRWKRNF 300
301 KKENPWLRDD LSIFGLWAYD STTALAMAVE KTNISSFPYN NASGSSNNMT DLGTLHVSRY 360
361 GPSLLEALSE IRFNGLAGRF NLIDRQLESP KFEIINFVGN EERIVGFWTP SNGLVNVNSN 420
421 KTTSFTGERF GPLIWPGKST IVPKGWEIPT NGKKIKVGVP VKKGFFNFVE VITDPITNIT 480
481 TPKGYAIDIF EAALKKLPYS VIPQYYRFES PDDDYDDLVY KVDNGTLDAV VGDVTITAYR 540
541 SLYADFTLPY TESGVSMMVP VRDNENKNTW VFLKPWGLDL WVTTACFFVL IGFVVWLFEH 600
601 RVNTDFRGPP HHQIGTSFWF SFSTMVFAHR EKVVSNLARF VVVVWCFVVL VLTQSYTANL 660
661 TSFLTVQRFQ PAAINVKDLI KNGDYVGYQH GAFVKDFLIK EGFNVSKLKP FGSSEECHAL 720
721 LSNGSISAAF DEVAYLRAIL SQYCSKYAIV EPTFKTAGFG FAFPRNSPLT GDVSKAILNV 780
781 TQGDEMQHIE NKWFMKQNDC PDPKTALSSN RLSLRSFWGL FLIAGIASFL ALLIFVFLFL 840
841 YENRHTLCDD SEDSIWRKLT SLFRNFDEKD IKSHTFKSSA VHHVSSPMTQ YIPSPSTLQI 900
901 APRPHSPSQD RAFELRRVSF TPNEERLTTQ TIHFEDEESD IECVVEQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
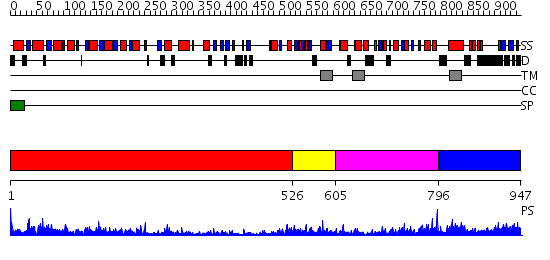
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..525] | 48.69897 | No description for 2e4uA was found. | |
| 2 | View Details | [526..604] | 2.29 | Structure of the Kainate Receptor Subunit GluR6 Agonist Binding Domain Complexed with Domoic Acid | |
| 3 | View Details | [605..795] | 21.0 | Glutamine-binding protein | |
| 4 | View Details | [796..947] | 2.491994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|