













| General Information: |
|
| Name(s) found: |
dlk-1 /
CE36155
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 577 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA synapse [IDA |
| Biological Process: |
activation of MAPKK activity
[IGI regulation of synapse organization [IGI signal transduction [IEA nematode larval development [IMP protein amino acid phosphorylation [IEA |
| Molecular Function: |
magnesium ion binding
[IEA ATP binding [IEA protein tyrosine kinase activity [IEA non-membrane spanning protein tyrosine kinase activity [IEA protein C-terminus binding [IPI protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSTTMVTTL DLVTPTSEEQ PGPAPESSDF STVVLSPDGS ELVTQSAPNT PIQHREQANA 60
61 EFGQKEGSPD PKNMVAATGN ASKPSLNSFY ADGLGQLRNG LFSCFQPVFG YFGTKTTVEI 120
121 EKSEDELWEI PFDAISELEW LGSGSQGAVF RGQLENRTVA VKKVNQLKET EIKHLRHLRH 180
181 QNIIEFLGVC SKSPCYCIVM EYCSKGQLCT VLKSRNTITR ELFAQWVKEI ADGMHYLHQN 240
241 KVIHRDLKSP NILISAEDSI KICDFGTSHM QKKMDSTMMS FCGTVSWMAP EMIKKQPCNE 300
301 KVDVYSFGVV LWEMLTRETP YANIAQMAII FGVGTNILSL PMPEEAPKGL VLLIKQCLSQ 360
361 KGRNRPSFSH IRQHWEIFKP ELFEMTEEEW QLAWDSYREF AKCIQYPSTV TRDHGGPKSA 420
421 FAMEEEIQRK RHEQLNHIKD IRNMYEMKLK RTNKMYDKLQ GCFTELKLKE SELAEWEKDL 480
481 TEREQWHNQN SPKAVAAPRA QLRGYPNEGY DDMSSDEDVQ PCRGSPYRCS NTSSSSGVQS 540
541 SPFSRQSSSR SSAGQQTRRS EGANPPSEYI FPEKLNI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
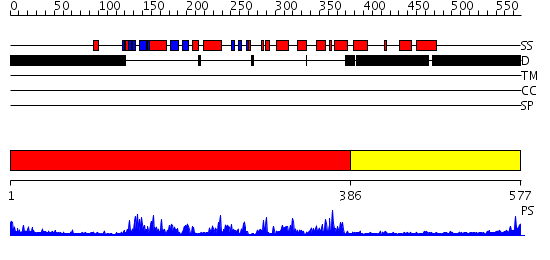
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..385] | 76.69897 | No description for 2fo0A was found. | |
| 2 | View Details | [386..577] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)