













| General Information: |
|
| Name(s) found: |
CE36099
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 397 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
lysine biosynthetic process
[IEA arginine biosynthetic process [IEA lysine biosynthetic process via diaminopimelate [IEA proteolysis [IEA metabolic process [IEA cellular amino acid metabolic process [IEA |
| Molecular Function: |
zinc ion binding
[IEA hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines [IEA metal ion binding [IEA dipeptidase activity [IEA protein dimerization activity [IEA succinyl-diaminopimelate desuccinylase activity [IEA aminoacylase activity [IEA acetylornithine deacetylase activity [IEA metallopeptidase activity [IEA cobalt ion binding [IEA hydrolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEEHIAVTR FREYLRVNTE QPNPNYAACR DFLFKYADEL GIARRSIETA PGVFLVIMTI 60
61 PGSQPELPSI MLYSHTDVVP TFREHWTHDP YSAFKDEDGN IFARGAQDMK CVGVQQMEAL 120
121 RNLFAQGIRQ WKRTIHLVWG PDEEIFGING MKGFAKTDEF KKLNLGFSLD EGMPSDDDVY 180
181 KVFYAERVAW WVKVTFPGNP GHGSQFMENT AMEKLERFLA SARKFRNEQK VVLESNPNLT 240
241 LGDVTTLNVN IVNGGVQFNV IPEKFEAYVD MRLTPHEDFN KIREMLDQWA KNAGEGVTYE 300
301 FSQYSDQKPI SAHTRDNSFW AAFEDSLNQE NCKFEKGIMV ASTDSRFVRY EGVNSINFSP 360
361 MINTPFLPHD HNEYLNEKVF LRGLEIYQTI INNLGNV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
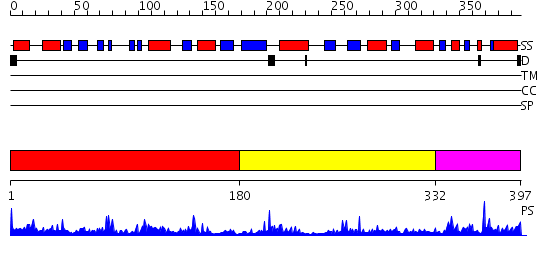
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] | 36.221849 | Zn-binding domain of the T347G mutant of human aminoacylase-I | |
| 2 | View Details | [180..331] | 4.07 | Crystal Structure of Bacillus Subtilis YXEP Protein (APC1829), a Dinuclear Metal Binding Peptidase from M20 Family | |
| 3 | View Details | [332..397] | 10.69897 | Zn-binding domain of the T347G mutant of human aminoacylase-I |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)