













| General Information: |
|
| Name(s) found: |
acs-9 /
CE35779
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 497 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA metabolic process [IEA phenylpropanoid metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA 4-coumarate-CoA ligase activity [IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKYYPETLF HDLILENVVK FGIRQALVHD NQVITFEEIP QLVSKLVYKL LELGISQGDT 60
61 ILVCLPNSIW YPLLFLSCAK IGAVLSGISH ESTAGEIKYS LKQSGAKLVF TNEKVSKENY 120
121 WALSENSVEV LPDSVKTYID RITGHEDFRP ENLDIDSILL APFSSGTTGA PKCCLLTHRN 180
181 FLAATYSLKK FLFDQLLAQS SMKTLAFLPF HHASGFWALL ICLLEGCTTY IMSEFHPIVM 240
241 MDLIEKYEID TINIVPPIAN IFLKMGILQG RCPSLRTILC GSSGLQKDRC KRLLSIFPQV 300
301 THFIQGYGMT ELVVLSCVTP FDDNFEHLGS CGHILPGFET KLFEHPTGET ELWLKSDAIM 360
361 KAYKNGTPNL DEDDWLHTGD IVTEKGGFFY VVDRMKDLIK LNGYQVSPTE IENVILTLPK 420
421 VAEVAVVGIE DELCGQLPKA YIVLEKNADE LLFLKHLEHT MKEKLSAVKQ LRGGVSIIKE 480
481 MPKSSSGKIQ KNRLMYY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
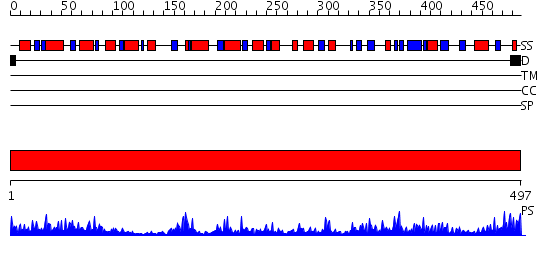
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..497] | 96.0 | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)