













| General Information: |
|
| Name(s) found: |
alh-1 /
CE32434
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IEA cytoplasm [IEA |
| Biological Process: |
cellular aldehyde metabolic process
[IEA metabolic process [IEA 10-formyltetrahydrofolate catabolic process [IEA proline biosynthetic process [IEA valine metabolic process [IEA betaine biosynthetic process [IEA one-carbon metabolic process [IEA |
| Molecular Function: |
1-pyrroline-5-carboxylate dehydrogenase activity
[IEA betaine-aldehyde dehydrogenase activity [IEA oxidoreductase activity [IEA aldehyde dehydrogenase [NAD(P)+] activity [IEA methylmalonate-semialdehyde dehydrogenase (acylating) activity [IEA formyltetrahydrofolate dehydrogenase activity [IEA oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDASQRGVLL NRLADLMERD RVILASLESL DNGKPYAVAY NADLPLSIKT LRYYAGWADK 60
61 NHGKTIPIEG DYFTYTRHEP VGVCGQIIPW NFPLLMQAWK LGPALAMGNT VVMKVAEQTP 120
121 LSALHVAALT KEAGFPDGVV NIIPGYGHTA GQAISSHMDV DKVAFTGSTE VGRLVMKAAA 180
181 ESNVKKVTLE LGGKSPNIIF ADADLNDSVH QANHGLFFNQ GQCCCAGSRT FVEGKIYDDF 240
241 VARSKELAEK AVIGDPFDLK TTQGPQVDGK QVETILKYIA AGKKDGAQLV TGGAKHGDQG 300
301 HFVKPTIFAN VKDQMTIAQE EIFGPVMTII RFDTMEELVE KANNTIYGLA AGVMTKDIDK 360
361 ALHIANATRA GSVWVNCYDV FDAAAPFGGF KQSGIGRELG EYGLEAYTEV KTVTIKVPQK 420
421 NS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
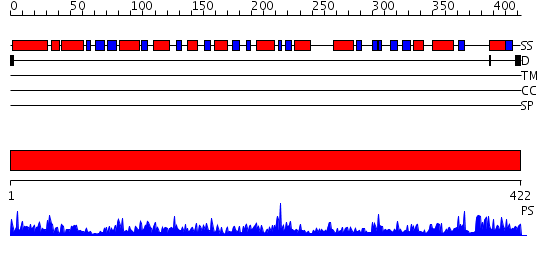
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..422] | 155.0 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)