













| General Information: |
|
| Name(s) found: |
alh-1 /
CE29809
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 510 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IEA cytoplasm [IEA |
| Biological Process: |
cellular aldehyde metabolic process
[IEA metabolic process [IEA 10-formyltetrahydrofolate catabolic process [IEA proline biosynthetic process [IEA valine metabolic process [IEA betaine biosynthetic process [IEA one-carbon metabolic process [IEA |
| Molecular Function: |
1-pyrroline-5-carboxylate dehydrogenase activity
[IEA betaine-aldehyde dehydrogenase activity [IEA oxidoreductase activity [IEA aldehyde dehydrogenase [NAD(P)+] activity [IEA methylmalonate-semialdehyde dehydrogenase (acylating) activity [IEA formyltetrahydrofolate dehydrogenase activity [IEA oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRSALRATV QARNASGVPP GLSNMKPQYT GIFINNEFVP AKSGKTFETI NPANGKVLAQ 60
61 VAEGDKTDVN IAVKAAQNAF RIGSEWRRMD ASQRGVLLNR LADLMERDRV ILASLESLDN 120
121 GKPYAVAYNA DLPLSIKTLR YYAGWADKNH GKTIPIEGDY FTYTRHEPVG VCGQIIPWNF 180
181 PLLMQAWKLG PALAMGNTVV MKVAEQTPLS ALHVAALTKE AGFPDGVVNI IPGYGHTAGQ 240
241 AISSHMDVDK VAFTGSTEVG RLVMKAAAES NVKKVTLELG GKSPNIIFAD ADLNDSVHQA 300
301 NHGLFFNQGQ CCCAGSRTFV EGKIYDDFVA RSKELAEKAV IGDPFDLKTT QGPQVDGKQV 360
361 ETILKYIAAG KKDGAQLVTG GAKHGDQGHF VKPTIFANVK DQMTIAQEEI FGPVMTIIRF 420
421 DTMEELVEKA NNTIYGLAAG VMTKDIDKAL HIANATRAGS VWVNCYDVFD AAAPFGGFKQ 480
481 SGIGRELGEY GLEAYTEVKT VTIKVPQKNS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
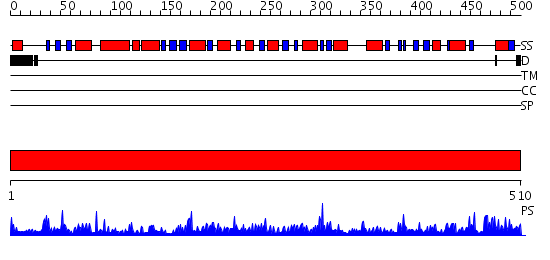
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..510] | 1000.0 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)