













| General Information: |
|
| Name(s) found: |
CE30606
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 690 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
chromatin modification
[IEA reproduction [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA nucleotide binding [IEA DNA binding [IEA ATP binding [IEA nucleoside-triphosphatase activity [IEA helicase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLTDEQRQA IAKKREEAIR RAAAYREREM QAAANATAST SSAAPPAPPK PVIPVMFSQN 60
61 RQNFQPMKPT MNNSTKQSTI NNYIKQVQKP EPTSLIKPTI GVKLKLDIGD RIKIEFYPFH 120
121 SAVIDLIKQV PSRNYDPAKR SWTVASSDHI TISNILKNAT AVKVELEPLP QNIIGLTNFK 180
181 PKAAPSDLNT VMDPSLIERL FPYQKEGVIF ALERDGRILL ADEMGLGKSV QALTIARYYK 240
241 ADWPLLIVCP ASVKGAWKKQ LNTFFPIIHR IFIVDKSSDP LPDVCTSNTV AIMSYEQMVL 300
301 KHDILKKEKY RTIIFDESHM LKDGKARRTK VATDLSKVAL HVILLSGTPA LSRPSELFTQ 360
361 IRLIDHKLFT NFHEFAIRYC DGKQGRFCFE AKGCTNSEEL AAIMFKRLMI RRLKADVLKD 420
421 LPEKRREVVY VSGPTIDARM DDLQKARADY EKVNSMERKH ESLLEFYSLT GIVKAAAVCE 480
481 HILENYFYPD APPRKVLIFA HHQIVLDTIQ VEVNKRKLGS IRIDGKTPSH RRTALCDSFQ 540
541 TDDNIRVAVL SITAAGVGIT LTAASVVVFA EIHFNPGYLV QAEDRAHRVG QKDSVFVQYL 600
601 IAKKTADDVM WNMVQQKLDV LGQVSLSSDT FRTADKMHLR FNDAAQPGIA EYLKKTPDTT 660
661 IDEWEDPVEE KEDDDLEIIC DSPAPKRIKN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
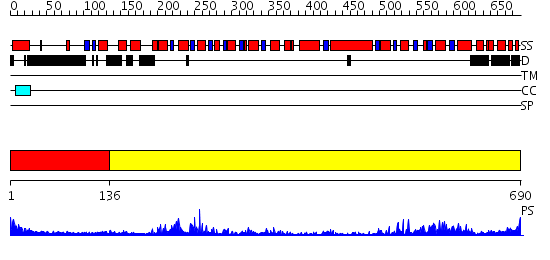
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [136..690] | 74.39794 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)