













| General Information: |
|
| Name(s) found: |
csn-1 /
CE26200
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic cleavage
[IMP establishment of mitotic spindle orientation [IMP embryonic development ending in birth or egg hatching [IMP cytokinesis [IMP gamete generation [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNELLDDPM DTGAPAAEAA AADEPLPPAH GIDREVTPKP AGNPDLMQQS LDNPAIPMHL 60
61 IEKRDDRRES CDDGYSLTVN ESAIDIESLA CSYDSNAFFL RARFIARHCP ILRADAYISL 120
121 INYLKEHTTD ITHYVAFFNE LESELARKEF KNRQLNFQIP LRDQKWIEEN GATWQSTTDQ 180
181 LQAEYKRHKD EGVKESTRRA MEDLFQHYMM AGKIDEAIRL YSRGIRDYCT QLKHSINMWI 240
241 NWMEVAICAN DWGKLDTVTN TAYRSLKDAD DAEKNSQQSQ QAPPQRGENA PYMVERDPNA 300
301 PIQTLTNRQL IETALAKCLA AQVLLKLRNK RYSQVVETIL QIKTECLQSK WFVTSSDLGI 360
361 YGMLSAMATM SRADLKLQVS GNGTFRKLLE SEPQLIELLG SYTSSRFGRC FEIMRSVKPR 420
421 LLLDPFISRN VDELFEKIRQ KCVLQYLQPY STIKMATMAE AVGMSSAELQ LSLLELIEQK 480
481 HVSLKIDQNE GIVRILDERD ENAILKRVNV TCDRATQKAK SLLWKTTLAG ANIHSISDKE 540
541 TRPKRKNQKE SAKFDRNFGG IDVDEDPRGI AGPSGLSDDF NIAYDQQPQQ QVQYLEDLGD 600
601 I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
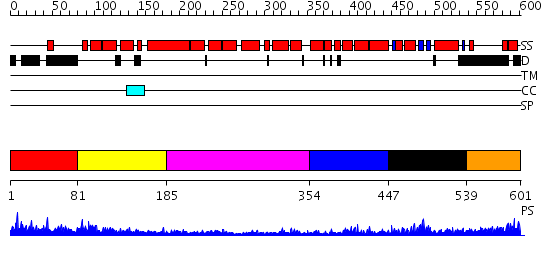
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..184] | 4.393997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [185..353] | 4.154902 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 4 | View Details | [354..446] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [447..538] | 9.0 | Solution structure of the PCI domain | |
| 6 | View Details | [539..601] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)