













| General Information: |
|
| Name(s) found: |
mec-18 /
CE16940
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell
[IDA |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA metabolic process [IEA menaquinone biosynthetic process [IEA detection of mechanical stimulus involved in sensory perception of touch [IMP siderophore biosynthetic process [IEA |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA CoA-ligase activity [TAS] AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIQNPFQIS WDKTAAQSFH DYFFSKISAH GSNLAIVDVD SGKQWRYSEI RNWCEMCATR 60
61 LKELQVTSGS RVAVITGTTG QAIFVHLACS IIGCSAVAVN GWNAVDEIWT LVDLSEATHL 120
121 IVENQFMQKA DDVRRKAQMR GGGRIKHVRQ IEDVLTAERI NVDSARKRSI DVKQKPPLVK 180
181 EMSLAKINLT PVLPEDIADL TSPVSEVSGQ SKELNGDVES SVVDSEEVQT PIQQVSGQNP 240
241 LLILFTSGTT GLAKAAELSH RSLIINIQQI SLPLYGPVQT KERFLLPLSI AHIYGIVSAY 300
301 YALINGASLY LISKQSNRLF METLVNNQIN VMHITPAIVH WMATDAIVDD YKTPNLRSVL 360
361 CAGAPIDSNS AAAMKSRLNI KDFRQSFGMT ELGGICTMSP YLDEKIESVG NPLPGMLFKV 420
421 VNWETKQLCL PRQPGQIIVL GPQVSPCYYK NPKATSELFD ATGFVKTGDA GFYDEVGRIY 480
481 VLDRIKDIIK CKGTMICPSE VELVLRAHAG IDDCAVVGRQ DHVTGEVPAA FVVKNAQHPL 540
541 LASAEVRQYV SGKIATFKEL RGGVFFISEI PRSVCGKILR RNLRQFWDRE RTNSKVDSLT 600
601 NKDGAKTAGA AKGAPAKKVN GSTTPSRRPP SAGRPVKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
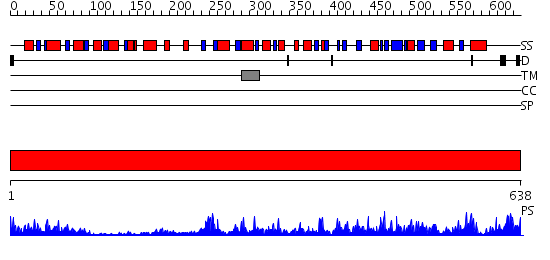
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..638] | 96.39794 | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.32 |
Source: Reynolds et al. (2008)