













| General Information: |
|
| Name(s) found: |
usp-14 /
CE15615
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 489 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein modification process
[IEA ubiquitin-dependent protein catabolic process [IEA |
| Molecular Function: |
ubiquitin thiolesterase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIVNVKWQK EKYVVEVDTS APPMVFKAQL FALTQVVPER QKVVIMGRTL GDDDWEGITI 60
61 KENMTIMMMG SVGEIPKPPT VLEKKQANRD KQAEEISALY PCGLANLGNT CYFNSCVQML 120
121 KEVNELVLKP AEEMRIREHN DRLCHNLATL FNSLRDKDRA LRSKGEPIKP FAAILTLSDS 180
181 FPQFEKFKQQ DANECLVSIM SNVTRIYGLS GWNIESLFRI QTETTMKCLE SDEVSEKKVE 240
241 RNNQLTCYVN QDVRFLQTGI KAGFEEEMTR NSEELNRDAK WQKNTQISRL PKYLTVNINR 300
301 FFYKESTKTN AKILKSVQFP MQLDTYDLCS QELKDKLVAR RADIKLEEDA KLERELRKKV 360
361 LDKEQGDKIF DDGVALPTAF EDDAGSNNSG FYDLKGIITH KGRSSQDGHY VAWMRSSEDG 420
421 KWRLFDDEHV TVVDEEAILK TSGGGDWHSA YVLLYEARVI KQFPELPPAP VPTEVAADTA 480
481 EPMEVSEKQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
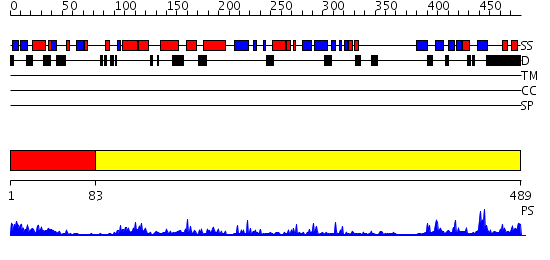
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 10.30103 | Solution Structure of the N-terminal Ubiquitin-like Domain of Mouse Ubiquitin Specific Protease 14 (USP14) | |
| 2 | View Details | [83..489] | 45.522879 | Structure of USP14, a proteasome-associated deubiquitinating enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)