













| General Information: |
|
| Name(s) found: |
acs-21 /
CE08917
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 517 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA metabolic process [IEA phenylpropanoid metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA 4-coumarate-CoA ligase activity [IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTHHLKSSI HTTAGLRAAT SAANNIISQA NLQSDISKVL FIDDGRKITY GDFVQRAGQY 60
61 ATALTEKYKI KKGDRVMARV SKTTDTAALY AACLQIGALY VPVNPALTQS EAAHYVKDAK 120
121 PSLWITCKDD ADQIAMFQRV LKTVCIDNPV EVINEKVLAD EAGKRKACTM IEHVEKSNAA 180
181 TICFTSGTTG APKGAVLSHG ALTNNTNALV QEWGFTENDV NLHCLPIFHA HGLYFSLHCS 240
241 LFSHSSVIWR PNFDAEDCSK HLKNATVFMG VPTFYSRLLA TNNFNKESFE KIRLFISGSA 300
301 PLSVPTLEEF EKRTGQVILE RYGMTEAGVI ASNPLKGKRK AGTVGQALKG VQCRVTENGE 360
361 IEIKSDSIFS EYWKNPEKTK EEFTEDGWFK TGDVGSLDKD GYLTIGGRSK DMIISGGENI 420
421 YPKEIEDAID SIEFVKESAV IAAPHPDFGE AVVAVVVPKN MVEDEQKFEE DLIEMLRKKL 480
481 AKYKVPKKVI LLEELPRNHI TKVRKDILRA SYNKLFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
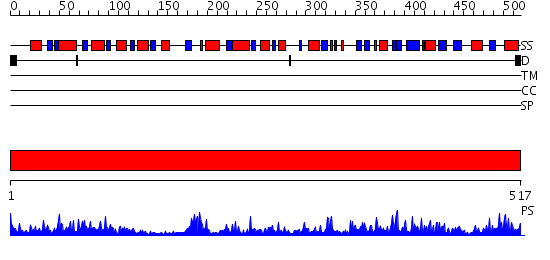
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..517] | 115.0 | Crystal structure of tt0168 from Thermus thermophilus HB8 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|