













| General Information: |
|
| Name(s) found: |
acs-6 /
CE07642
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 565 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVFVSKAASV KLSTTTVHER IFEACRIHAS ANKDAVCFID AETTTKKKLY KDVEPTVNSL 60
61 ATALVKLGFK PGDVASQAFP NCPEFLVAML AVMKCGGAMS NASAIFTDYE LQLQFCDSNT 120
121 SIVFTDEDRL ARIRRATAKC PGVRKIICLR TFPLRTEFPE NVLDYVELTQ TPDQPINVNV 180
181 SMDSIALLPY SSGTTGRPKG CQLTHRNIGA MLDVAKAHLE TDVAPAMFGK EKATWHKEHT 240
241 VLLLPWYHAY GLNTMFETIL LGMTGIVFKK FDTIVMLNRI KFYKVKLAWL VPPMLIFLAK 300
301 DPMVPIFNTA PFLKVIMSAG ATAGKQLCEE VSKRFPNAWL CQAYGMTEMV QFTTIPRFED 360
361 GNCFETVGNL ASTYELKILD KEKKEITTIN TVGQLCFRGP TVMKGYLKRE EADIIDKDGF 420
421 LLTGDLGSID DKGRIHVTGR IKELIKVNGM QVPPVEIEDV LLLHPKVKDC AVIGVPDEHK 480
481 GESPKAYIVK KDHTLTEAEL TEFVRQKLSS YKWIDTYEFI DSIPKLPSGK IQRKKLKKMA 540
541 ESSGSTESEA SEPSQSERSS HSIRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
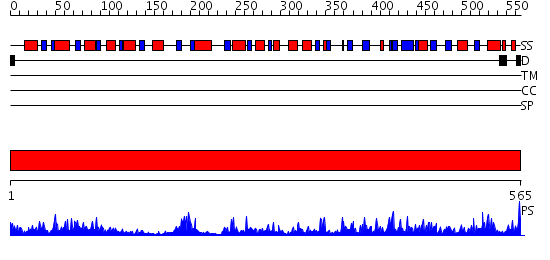
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..565] | 109.0 | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)